



Next: Free Energy of Conformational
Up: Additional Simulation Parameters
Previous: Pressure Control
Contents
Index
Subsections
There are several ways to apply external forces to simulations with NAMD.
These are described below.
NAMD provides the ability to apply constant forces to some atoms.
There are two parameters that control this feature.
NAMD provides the ability to apply a constant electric field to the molecular
system being simulated.
Energy due to the external field will be reported in the MISC column
and may be discontinuous in simulations using periodic boundary conditions if,
for example, a charged hydrogen group moves outside of the central cell.
There are two parameters that control this feature.
Moving constraints feature works in conjunction with the Harmonic
Constraints (see an appropriate section of the User's guide).
The reference positions of all constraints
will move according to
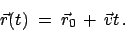 |
(1) |
A velocity vector 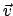 (movingConsVel) needs to be specified.
(movingConsVel) needs to be specified.
The way the moving constraints work is that the moving reference
position is calculated every integration time step using
Eq. 1, where 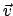 is in Å/timestep, and
is in Å/timestep, and 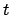 is the
current timestep (i.e., firstTimestep plus however many
timesteps have passed since the beginning of NAMD run). Therefore,
one should be careful when restarting simulations to appropriately
update the firstTimestep parameter in the NAMD configuration
file or the reference position specified in the reference PDB file.
is the
current timestep (i.e., firstTimestep plus however many
timesteps have passed since the beginning of NAMD run). Therefore,
one should be careful when restarting simulations to appropriately
update the firstTimestep parameter in the NAMD configuration
file or the reference position specified in the reference PDB file.
NOTE: NAMD actually calculates the constraints
potential with
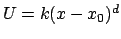 and the force with
and the force with
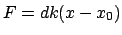 ,
where
,
where 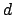 is the exponent consexp. The result is that if one
specifies some value for the force constant
is the exponent consexp. The result is that if one
specifies some value for the force constant 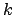 in the PDB file,
effectively, the force constant is
in the PDB file,
effectively, the force constant is 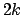 in calculations. This caveat
was removed in SMD feature.
in calculations. This caveat
was removed in SMD feature.
The following parameters describe the parameters for the
moving harmonic constraint feature of NAMD.
The constraints parameters are specified in the same manner as for
usual (static) harmonic constraints. The reference positions of all
constrained atoms are then rotated with a given angular velocity
about a given axis. If the force constant of the constraints is
sufficiently
large, the constrained atoms will follow their reference positions.
A rotation matrix 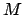 about the axis unit vector
about the axis unit vector 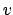 is calculated every
timestep
for the angle of rotation corresponding to the current timestep.
angle =
is calculated every
timestep
for the angle of rotation corresponding to the current timestep.
angle = 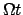 ,
where
,
where 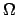 is the angular velocity of rotation.
is the angular velocity of rotation.
From now on, all quantities are 3D vectors, except the matrix 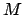 and the
force constant
and the
force constant 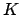 .
.
The current reference position 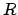 is calculated from the initial
reference
position
is calculated from the initial
reference
position 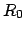 (at
(at 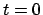 ),
),
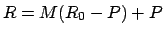 ,
where
,
where 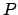 is the pivot point.
is the pivot point.
Coordinates of point N can be found as
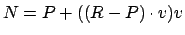 .
Normal from the atom pos to the axis is, similarly,
normal
.
Normal from the atom pos to the axis is, similarly,
normal
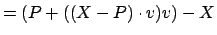 The force is, as usual,
The force is, as usual,
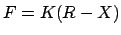 ;
This is the force applied to the atom in NAMD (see below).
NAMD does not know anything about the torque
applied. However, the torque applied to the atom can be calculated
as a vector product
torque
;
This is the force applied to the atom in NAMD (see below).
NAMD does not know anything about the torque
applied. However, the torque applied to the atom can be calculated
as a vector product
torque
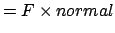 Finally, the torque applied to the atom with respect to the axis
is the projection of the torque on the axis, i.e.,
Finally, the torque applied to the atom with respect to the axis
is the projection of the torque on the axis, i.e.,
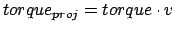
If there are atoms that have to be constrained, but not moved,
this implementation is not suitable, because it will move all
reference positions.
Only one of the moving and rotating constraints can be used at a
time.
Using very soft springs for rotating constraints leads to the system
lagging behind the reference positions, and then the force is applied
along a direction different from the "ideal" direction along the
circular path.
Pulling on N atoms at the same time with a spring of stiffness K
amounts to pulling on the whole system by a spring of stiffness NK,
so the overall behavior of the system is as if you are pulling with a
very stiff spring if N is large.
In both moving and rotating constraints the force constant that you
specify in the constraints pdb file is multiplied by 2 for the force
calculation, i.e., if you specified
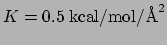 in the pdb
file,
the force actually calculated is
in the pdb
file,
the force actually calculated is
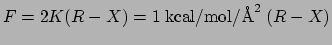 .
SMD feature of namd2 does the calculation without multiplication of
the
force constant specified in the config file by 2.
.
SMD feature of namd2 does the calculation without multiplication of
the
force constant specified in the config file by 2.
In TMD, subset of atoms in the simulation is guided towards a
final 'target' structure by means of steering forces. At each timestep,
the RMS distance between
the current coordinates and the target structure is computed (after
first aligning the target structure to the current coordinates).
The force on each atom is given by the gradient of the potential
![\begin{displaymath}
U_{TMD} = \frac{1}{2} \frac{k}{N} \left[ RMS(t) - RMS^*(t) \right]^2
\end{displaymath}](img72.png) |
(2) |
where 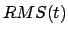 is the instantaneous best-fit RMS distance of the current
coordinates from the target coordinates, and
is the instantaneous best-fit RMS distance of the current
coordinates from the target coordinates, and 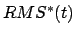 evolves linearly
from the initial RMSD at the first TMD step to the final RMSD at the last
TMD step. The spring constant
evolves linearly
from the initial RMSD at the first TMD step to the final RMSD at the last
TMD step. The spring constant 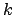 is scaled down by the number
is scaled down by the number 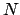 of targeted
atoms.
of targeted
atoms.
- TMD
 Is TMD active
Is TMD active 
Acceptable Values: on or off
Default Value: off
Description: Should TMD steering forces be applied to the system. If TMD is enabled,
TMDk, TMDFile, and TMDLastStep must be defined in the
input file as well.
- TMDk
 Elastic constant for TMD forces
Elastic constant for TMD forces 
Acceptable Values: Positive value in 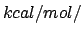 Å
Å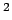 .
.
Description: The value of 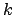 in Eq. 2. A value of 200 seems to work
well in many cases.
in Eq. 2. A value of 200 seems to work
well in many cases.
- TMDOutputFreq
 How often to print TMD output
How often to print TMD output 
Acceptable Values: Positive integer
Default Value: 1
Description: TMD output consists of lines of the form TMD ts targetRMS currentRMS
where ts is the timestep, targetRMS is the target RMSD at that
timestep, and currentRMS is the actual RMSD.
- TMDFile
 File for TMD information
File for TMD information 
Acceptable Values: Path to PDB file
Description:
Target atoms are those whose occupancy (O) is nonzero in the TMD PDB file.
The file must contain the same number of atoms as the structure file. The
coordinates for the target structure are also taken from the targeted
atoms in this file. Non-targeted atoms are ignored.
- TMDFirstStep
 first TMD timestep
first TMD timestep 
Acceptable Values: Positive integer
Default Value: 0
Description:
- TMDLastStep
 last TMD timestep
last TMD timestep 
Acceptable Values: Positive integer
Description: TMD forces are applied only between TMDFirstStep and TMDLastStep.
The target RMSD evolves linearly in time from the initial to the final target
value.
- TMDInitialRMSD
 target RMSD at first TMD step
target RMSD at first TMD step 
Acceptable Values: Non-negative value in Å
Default Value: from coordinates
Description:
In order to perform TMD calculations that involve restarting a previous
NAMD run, be sure to specify TMDInitialRMSD with the same value
in each NAMD input file, and use the NAMD parameter firstTimestep
in the continuation runs so that the target RMSD continues from where the
last run left off.
- TMDFinalRMSD
 target RMSD at last TMD step
target RMSD at last TMD step 
Acceptable Values: Non-negative value in Å
Default Value: 0
Description: If no TMDInitialRMSD is given, the initial RMSD will be calculated at the
first TMD step. TMDFinalRMSD may be less than or greater than
TMDInitialRMSD, depending on whether the system is to be steered
towards or away from a target structure, respectively. Forces are applied
only if 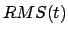 is betwween TMDInitialRMSD and
is betwween TMDInitialRMSD and 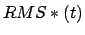 ; in other
words, only if the current RMSD fails to keep pace with the target value.
; in other
words, only if the current RMSD fails to keep pace with the target value.
The SMD feature is independent from the harmonic constraints, although it
follows the same ideas. In both SMD and harmonic constraints, one specifies
a PDB file which indicates which atoms are 'tagged' as constrained. The PDB
file also gives initial coordinates for the constraint positions. One also
specifies such parameters as the force constant(s) for the constraints,
and the velocity with which the constraints move.
There are two major differences between SMD and
harmonic constraints:
- In harmonic constraints, each tagged atom is harmonically constrained
to a reference point which moves with constant velocity. In SMD, it is
the center of mass of the tagged atoms which is constrained to move
with constant velocity.
- In harmonic constraints, each tagged atom is constrained in all three
spatial dimensions. In SMD, tagged atoms are constrained only along
the constraint direction.
The center of mass of the SMD atoms will be harmonically constrained with
force constant 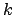 (SMDk) to move with velocity
(SMDk) to move with velocity 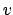 (SMDVel) in
the direction
(SMDVel) in
the direction 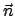 (SMDDir). SMD thus results in the following
potential being applied to the system:
(SMDDir). SMD thus results in the following
potential being applied to the system:
![\begin{displaymath}
U(\vec r_1, \vec r_2, ..., t) \; = \; \frac{1}{2}
k\left[vt - (\vec R(t) - \vec R_0)\cdot \vec n \right]^2.
\end{displaymath}](img80.png) |
(3) |
Here,
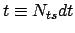 where
where 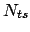 is the number of elapsed timesteps
in the simulation and
is the number of elapsed timesteps
in the simulation and 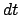 is the size of the timestep in femtoseconds.
Also,
is the size of the timestep in femtoseconds.
Also, 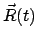 is the current center of mass of the SMD atoms and
is the current center of mass of the SMD atoms and 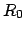 is
the initial center of mass as defined by the coordinates in SMDFile.
Vector
is
the initial center of mass as defined by the coordinates in SMDFile.
Vector 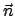 is normalized by NAMD before being used.
is normalized by NAMD before being used.
NAMD provides output of the current SMD data. The frequency of
output is specified by the SMDOutputFreq parameter in the
configuration file. Every SMDOutputFreq timesteps NAMD will
print the current timestep, current position of the center of mass of the
restrained atoms, and
the current force applied to the center of mass (in piconewtons, pN).
The output line starts with word SMD
The following parameters describe the parameters for the
SMD feature of NAMD.
NAMD now works directly with VMD to allow you to view and interactively
steer your simulation. With IMD enabled, you can connect to NAMD at any
time during the simulation to view the current state of the system or perform
interactive steering.
NAMD provides a limited Tcl scripting interface designed for applying forces and performing on-the-fly analysis.
This interface is efficient if only a few coordinates, either of individual atoms or centers of mass of groups of atoms, are needed.
In addition, information must be requested one timestep in advance.
To apply forces individually to a potentially large number of atoms, use
tclBC instead as described in Sec. 6.6.9.
The following configuration parameters are used to enable the Tcl interface:
At this point only low-level commands are defined.
In the future this list will be expanded. Current commands are:
- print <anything>
This command should be used instead of puts to display output.
For example, ``print Hello World''.
- atomid <segname> <resid> <atomname>
Determines atomid of an atom from its segment, residue, and name.
For example, ``atomid br 2 N''.
- addatom <atomid>
Request coordinates of this atom for next force evaluation, and
the calculated total force on this atom for current force evaluation.
Request remains in effect until clearconfig is called.
For example, ``addatom 4'' or ``addatom [atomid br 2 N]''.
- addgroup <atomid list>
Request center of mass coordinates of this group for next force evaluation.
Returns a group ID which is of the form gN where N is a small integer.
This group ID may then be used to find coordinates and apply forces just like a regular atom ID.
Aggregate forces may then be applied to the group as whole.
Request remains in effect until clearconfig is called.
For example, ``set groupid [addgroup { 14 10 12 }]''.
- clearconfig
Clears the current list of requested atoms. After clearconfig,
calls to addatom and addgroup can be used to build a new
configuration.
- getstep
Returns the current step number.
- loadcoords <varname>
Loads requested atom and group coordinates (in Å) into a local array.
loadcoords should only be called from within the calcforces procedure.
For example, ``loadcoords p'' and ``print $p(4)''.
- loadforces <varname>
Loads the forces applied in the previous timestep (in kcal mol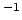 Å
Å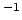 ) into a local array.
loadforces should only be called from within the calcforces procedure.
For example, ``loadforces f'' and ``print $f(4)''.
) into a local array.
loadforces should only be called from within the calcforces procedure.
For example, ``loadforces f'' and ``print $f(4)''.
- loadtotalforces <varname>
Loads the total forces on each requested atom in the previous
time step (in kcal mol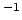 Å
Å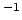 ) into a local array. The
total force also includes external forces. Note that the
``loadforces'' command returns external forces applied by
the user. Therefore, one can subtract the external force on an
atom from the total force on this atom to get the pure force
arising from the simulation system.
) into a local array. The
total force also includes external forces. Note that the
``loadforces'' command returns external forces applied by
the user. Therefore, one can subtract the external force on an
atom from the total force on this atom to get the pure force
arising from the simulation system.
- loadmasses <varname>
Loads requested atom and group masses (in amu) into a local array.
loadmasses should only be called from within the calcforces procedure.
For example, ``loadcoords m'' and ``print $m(4)''.
- addforce <atomid|groupid> <force vector>
Applies force (in kcal mol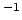 Å
Å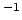 ) to atom or group.
addforce should only be called from within the calcforces procedure.
For example, ``
) to atom or group.
addforce should only be called from within the calcforces procedure.
For example, ``addforce $groupid { 1. 0. 2. }''.
- addenergy <energy (kcal/mol)>
This command adds the specified energy to the MISC column (and
hence the total energy) in the energy output. For normal runs,
the command does not affect the simulation trajectory at all, and
only has an artificial effect on its energy output. However, it
can indeed affect minimizations.
With the commands above and the functionality of the Tcl language,
one should be able to perform any on-the-fly analysis and
manipulation. To make it easier to perform certain tasks,
some Tcl routines are provided below.
Several vector routines (vecadd, vecsub, vecscale)
from the VMD Tcl interface are defined. Please refer to VMD
manual for their usage.
The following routines take atom coordinates as input, and return
some geometry parameters (bond, angle, dihedral).
- getbond <coor1> <coor2>
Returns the length of the bond between the two atoms. Actually
the return value is simply the distance between the two
coordinates. ``coor1'' and ``coor2'' are coordinates of the atoms.
- getangle <coor1> <coor2> <coor3>
Returns the angle (from 0 to 180) defined by the
three atoms. ``coor1'', ``coor2'' and ``coor3'' are coordinates of
the atoms.
- getdihedral <coor1> <coor2> <coor3> <coor4>
Returns the dihedral (from -180 to 180) defined by the
four atoms. ``coor1'', ``coor2'', ``coor3'' and ``coor4'' are
coordinates of the atoms.
The following routines calculate the derivatives (gradients) of
some geometry parameters (angle, dihedral).
As an example, here's a script which applies a harmonic
constraint (reference position being 0) to a dihedral. Note that
the ``addenergy'' line is not really necessary - it simply adds
the calculated constraining energy to the MISC column, which is
displayed in the energy output.
tclForcesScript {
# The IDs of the four atoms defining the dihedral
set aid1 112
set aid2 123
set aid3 117
set aid4 115
# The "spring constant" for the harmonic constraint
set k 3.0
addatom $aid1
addatom $aid2
addatom $aid3
addatom $aid4
set PI 3.1416
proc calcforces {} {
global aid1 aid2 aid3 aid4 k PI
loadcoords p
# Calculate the current dihedral
set phi [getdihedral $p($aid1) $p($aid2) $p($aid3) $p($aid4)]
# Change to radian
set phi [expr $phi*$PI/180]
# (optional) Add this constraining energy to "MISC" in the energy output
addenergy [expr $k*$phi*$phi/2.0]
# Calculate the "force" along the dihedral according to the harmonic constraint
set force [expr -$k*$phi]
# Calculate the gradients
foreach {g1 g2 g3 g4} [dihedralgrad $p($aid1) $p($aid2) $p($aid3) $p($aid4)] {}
# The force to be applied on each atom is proportional to its
# corresponding gradient
addforce $aid1 [vecscale $g1 $force]
addforce $aid2 [vecscale $g2 $force]
addforce $aid3 [vecscale $g3 $force]
addforce $aid4 [vecscale $g4 $force]
}
}
Tcl Boundary Forces
While the tclForces interface described above is very flexible, it is only
efficient for applying forces to a small number of pre-selected atoms.
Applying forces individually to a potentially large number of atoms, such
as applying boundary conditions, is much more efficient with the tclBC
facility described below.
The script provided in tclBCScript and the calcforces procedure
it defines are executed in multiple Tcl interpreters, one for every
processor that owns patches.
These tclBC interpreters do not share state with the Tcl interpreter used
for tclForces or config file parsing.
The calcforces procedure is passed as arguments
the current timestep, a ``unique'' flag which is non-zero for exactly
one Tcl interpreter in the simulation (that on the processor of patch zero),
and any arguments provided to the most recent tclBCArgs option.
The ``unique'' flag is useful to limit printing of messages, since the
command is invoked on multiple processors.
The print, vecadd, vecsub, vecscale,
getbond, getangle, getdihedral,
anglegrad, and dihedralgrad commands described under
tclForces are available at all times.
The wrapmode <mode> command, available in the tclBCScript or the
calcforces procedure, determines how coordinates obtained in the
calcforces procedure are wrapped around periodic boundaries. The options
are:
- patch, (default) the position in NAMD's internal patch data structure,
requires no extra calculation and is almost the same as cell
- input, the position corresponding to the input files of the simulation
- cell, the equivalent position in the unit cell centered on the cellOrigin
- nearest, the equivalent position nearest to the cellOrigin
The following commands are available from within the calcforces procedure:
- nextatom
Sets the internal counter to a new atom and return 1, or return 0
if all atoms have been processed (this may even happen the first call).
This should be called as the condition of a while loop, i.e.,
while {[nextatom]} { ... } to iterate over all atoms.
One one atom may be accessed at a time.
- dropatom
Excludes the current atom from future iterations on this processor
until cleardrops is called. Use this to eliminate extra work
when an atom will not be needed for future force calculations.
If the atom migrates to another processor it may reappear, so this
call should be used only as an optimization.
- cleardrops
All available atoms will be iterated over by nextatom as if
dropatom had never been called.
- getcoord
Returns a list {x y z} of the position of the current atom wrapped
in the periodic cell (if there is one) in the current wrapping mode
as specified by wrapmode.
- getcell
Returns a list of 1-4 vectors containing the cell origin (center)
and as many basis vectors as exist, i.e.,
{{ox oy oz} {ax ay az} {bx by bz} {cx cy cz}}.
It is more efficient to set the wrapping mode than to do periodic image
calculations in Tcl.
- getmass
Returns the mass of the current atom.
- getcharge
Returns the charge of the current atom.
- getid
Returns the 1-based ID of the current atom.
- addforce {<fx> <fy> <fz>}
Adds the specified force to the current atom for this step.
- addenergy <energy>
Adds potential energy to the BOUNDARY column of NAMD output.
As an example, these spherical boundary condition forces:
sphericalBC on
sphericalBCcenter 0.0,0.0,0.0
sphericalBCr1 48
sphericalBCk1 10
sphericalBCexp1 2
Are replicated in the following script:
tclBC on
tclBCScript {
proc veclen2 {v1} {
foreach {x1 y1 z1} $v1 { break }
return [expr $x1*$x1 + $y1*$y1 + $z1*$z1]
}
# wrapmode input
# wrapmode cell
# wrapmode nearest
# wrapmode patch ;# the default
proc calcforces {step unique R K} {
if { $step % 20 == 0 } {
cleardrops
# if $unique { print "clearing dropped atom list at step $step" }
}
set R [expr 1.*$R]
set R2 [expr $R*$R]
set tol 2.0
set cut2 [expr ($R-$tol)*($R-$tol)]
while {[nextatom]} {
# addenergy 1 ; # monitor how many atoms are checked
set rvec [getcoord]
set r2 [veclen2 $rvec]
if { $r2 < $cut2 } {
dropatom
continue
}
if { $r2 > $R2 } {
# addenergy 1 ; # monitor how many atoms are affected
set r [expr sqrt($r2)]
addenergy [expr $K*($r - $R)*($r - $R)]
addforce [vecscale $rvec [expr -2.*$K*($r-$R)/$r]]
}
}
}
}
tclBCArgs {48.0 10.0}




Next: Free Energy of Conformational
Up: Additional Simulation Parameters
Previous: Pressure Control
Contents
Index
namd@ks.uiuc.edu