


Next: Residue Selection
Up: Aquaporin Tutorial
Previous: Structural Alignment of Aquaporins
Contents
Subsections
Comparing Protein Sequence and Structure
In this unit you will study the structure and sequence conservation of the different
aquaporins you have aligned in the Multiseq window. Conservation is a term we will use frequently in exercises
from this point forward. Within the context of protein analysis, conservation refers to high levels
of similarity contingent on the employed metric. Structure conservation occurs when the structural
aspects (e.g. 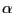 helix,
helix, 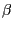 sheets), of the aligned proteins are highly similar. Likewise,
sequence conservation happens when at certain points of the aligned protein molecules the amino acid of each molecule is the same.
sheets), of the aligned proteins are highly similar. Likewise,
sequence conservation happens when at certain points of the aligned protein molecules the amino acid of each molecule is the same.
In order to better understand the structural conservation within the aligned
molecules, Multiseq provides tools that allow you to color the molecules according
to their Q value (Qres), a measure of structural conservation.
You will now color the molecules according to the value of Q per residue
obtained in the alignment.
- 1
- In the Multiseq program window, choose the View
 Coloring
Coloring  Qres (Fig. 13).
Qres (Fig. 13).
Figure:
Molecule Coloring
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.8 in]{pictures/colormenu}
}
\end{center}\vspace{-0.7cm}\end{figure}](img33.gif) |
- 2
- Look at the OpenGL window to see the impact this selection has
made on the coloring of the aligned molecules (Figure 14).
Figure:
Molecules colored by Qres value
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.5 in]{pictures/colorbyq}
}
\end{center}\vspace{-0.7cm}\end{figure}](img34.gif) |
Rotate the molecule to see how much of it has turned blue. Notice that the transmembrane helices of the aligned molecules have
turned blue. The blue areas indicate that the molecules are structurally conserved at those
points. If there is no correspondence in structural proximities at these
points, the points appear red. Observe how the structurally least similar segments
tend to be on the periphery of the aligned molecule. Note that the loops tend
to be red, while the helices are blue.
You have examined the structural similarity between the molecules. Now
you will look at the sequence conservation.
Multiseq has a feature to color the molecules according to how much the
sequence is conserved within the aligned molecules. This tool, Sequence
identity, colors each amino acid according to the degree of conservation
within the alignment: blue means highly conserved, whereas red means very low or no conservation.
- 1
- Choose View
 Coloring
Coloring
 Sequence Identity.
Sequence Identity.
Before you look at the viewer window, can you anticipate what will happen
to the coloring of the molecules? Will the molecules still be blue in the transmembrane region, as they
were when Qres was used to determine structure conservation?
- 2
- Now take a look at the viewer window. As you can see (Fig. 15), a fair portion of the molecules has
turned red, indicating less sequence conservation than structural similarity.
Figure:
Molecules colored by sequence identity
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.3 in]{pictures/colorbyseqid}
}
\end{center}\vspace{-0.7cm}\end{figure}](img35.gif) |
- 3
- Note that the residues at the site where the two short helices interact
are blue. Look at the molecules from the top (c.f. Fig. 16). Do you notice the blue residues
tend to be on the inside of the pore?
Figure:
Conserved residues (shown in blue) are located inside the
pore.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.5 in]{pictures/top}
}
\end{center}\vspace{-0.7cm}\end{figure}](img36.gif) |
Note that the coloring of the molecules using Sequence Identity
indicates that the sequence conservation is much less in comparison to the
structural conservation. Sometimes structures from the same family have less than
10% sequence identity, yet are structurally similar. A well known example is
the protein myoglobin that we recommend for self-study.
To examine the relationship between sequence and structure in more
detail, in the next section you will use the Select Residues feature.



Next: Residue Selection
Up: Aquaporin Tutorial
Previous: Structural Alignment of Aquaporins
Contents
school@ks.uiuc.edu
