In this part of the tutorial you will construct an energy gap
function through the molecular dynamics
study of an electron transport protein.
The computational demands of this section
of the tutorial are rather large compared to what you have
studied in previous tutorials.
You may want to
immediately get started on your first molecular
dynamics run of the day and then read on further
while your simulation is running.
Switch to the directory required for this
section by typing
tbss> cd ![]() /tbss.work/photo-tutorial-files/2-electron
/tbss.work/photo-tutorial-files/2-electron
This will henceforth be referred to as your working directory.
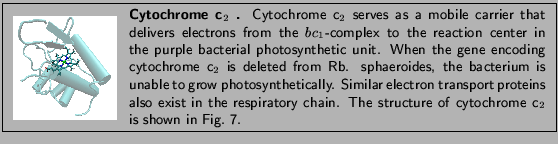
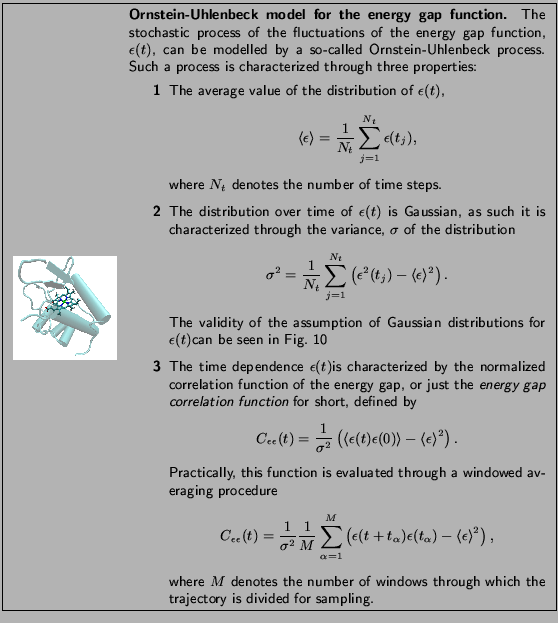
The system you are currently
simulating is the electron transport
protein cytochrome c![]() from the
purple bacterium Rhodobacter sphaeroides.
You have three main steps to perform in this section:
from the
purple bacterium Rhodobacter sphaeroides.
You have three main steps to perform in this section:
After starting your simulation,
let us first take a closer look at
cytochrome c![]() .
.
To explore the structure of cytochrome c![]() we will use VMD. Start VMD and load
the required coordinate and parameter files
by typing
we will use VMD. Start VMD and load
the required coordinate and parameter files
by typing
tbss> vmd -e cytexamine.vmd
in your working directory. This loads VMD
with a state file that allows you to view
the equilibrated
cytochrome c![]() structure in the beginning of the
simulation. (You do not need to type these lines
as they are taken from the state
file you have just loaded.)
structure in the beginning of the
simulation. (You do not need to type these lines
as they are taken from the state
file you have just loaded.)
![]()
mol new cyt_reduced.psf type psf
mol addfile SPH_1CXCequi.pdb type pdb
mol representation Cartoon 2.100000 12.000000 5.000000
mol color ColorID 7
mol selection protein
![]()
mol representation Bonds 0.300000 6.000000
mol color Name
mol selection resname HEMP
![]()
This should give you a picture that resembles Fig. 7. You may also turn on the water molecules to note that this system is solvated in a water box. Indeed your current simulation has started from an already minimized and equilibrated state with periodic boundary conditions. Let us now take a closer look at the simulation configuration you are currently running.

The NAMD configuration file cytreduced.namd
you executed earlier continues your simulation
from a restart point.
![]()
set inputname cyt_red_init
bincoordinates $inputname.rst.coor
ExtendedSystem $inputname.rst.xsc
binvelocities $inputname.rst.vel
![]()
Without a restart point you
would have had to worry about
velocity relaxations and discard the initial part
of your already short simulation.
Closer examination of the configuration file
will reveal use of periodic boundary
conditions.
Also of interest is the unusual
and ordinarily costly fact that you are
writing a DCD frame at each
and every time step.
![]()
outputEnergies 1
dcdfreq 1
![]()
This is needed to be able to probe the time
dependence of the energy gap function you
will compute below.
The output of the first namd run will be written to a DCD file called cytreduced.dcd, which in turn will be read by a second NAMD run described below.
Now you will start a new simulation to read the trajectory created by the first one to compute the interation between two sets of atoms in your simulation. In section 2.1.5 of the NAMD tutorial of past week, you have seen how to compute the specific heat of a protein by a similar technique.

The configuration file you just executed, cytrun2.namd, refers to a new structure file, cyt_deltaQ.psf, which contains the information about the charge differences between the reduced and oxidized forms of the HEME group. Hence, the total electrostatic energy computed by this run, appearing in the ELECT column of the output file, will correspond to the energy gap function defined above.
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
... similar method was employed for
specific heat computations.
}
\end{minipage} }](img64.gif)
In order to extract the gap function
information, after your second NAMD run is over, type:
tbss> namddat ELECT cytrun2.out
This writes the gap function to the file data.dat.
Now cut the header (i.e. the first line) of the output data.dat
with an editor, so we can read it
easily with a script. For example, you may use pico
tbss> pico data.dat
Delete the first line TS ELECT so that the file consists only of two columns of numbers.
As in the previous part of the tutorial you will
use Mathematica to display your results.
In the limited time frame of a tutorial session,
it is not possible to produce and analyze a long
molecular dynamics trajectory. For your
convenience, a longer 10 ps version of the energy gap function
data file, dataLONG.dat, is provided for you.
In order to use Mathematica to extract
the energy gap function from the data
file. First type
tbss> mathematica energygap.nb &
and then select Kernel ![]() Evaluation
Evaluation ![]() Evaluate Notebook (See Fig. 8.). This
will read the data file containing the energy
gap function you have created earlier,
as well as a longer data file containing
the data from a 10 ps simulation.
You can now examine your data more
closely. In particular, note that
the histogram of the energy gap
function over time gives a Gaussian
distribution. This concludes the last
section of this tutorial.
Evaluate Notebook (See Fig. 8.). This
will read the data file containing the energy
gap function you have created earlier,
as well as a longer data file containing
the data from a 10 ps simulation.
You can now examine your data more
closely. In particular, note that
the histogram of the energy gap
function over time gives a Gaussian
distribution. This concludes the last
section of this tutorial.
![\begin{figure}\begin{center}
\par\par\latex{
\includegraphics[scale=0.7]{pictures/gapplot}
}
\end{center} \end{figure}](img68.gif) |