In this part of the tutorial, you will learn about the transfer of excitation energy between pairs of bacteriochlorophyl molecules (BChls) in light harvesting complex II (LH-II). The tutorial consists of two sections. In the first section, you will use VMD to investigate the structure of LH-II in order to see how proteins and pigments are organized in the complex. Then, in the second section you will calculate the rate of excitation energy transfer between pairs of BChls in LH-II using VMD and Mathematica.
The working directory for this tutorial is at
photo-tutorial-files/1-excitation
It contains five files and one directory.
1. lh2.pdb is a pdb file for LH-II.
2. TwoDimer.vmd is a VMD state file for two LH-II structural units.
3. EightDimer.vmd is a VMD state file for LH-II ring.
4. lh2-dipole.vmd is a VMD state file for showing direction of transition dipole moments of BChls in LH-II.
5. transfer.nb is a Mathematica file for excitation transfer rate calculation.
6. example-output is a directory containing example output
files e.g. OneDimer.vmd is a VMD state file for an LH-II
structural unit.
In this section, you are going to view and
investigate the three dimensional structure of the light
harvesting complex II from Rhodospirillum (Rh.) molischianum using
VMD.
Your objective in this section is to see how proteins and
pigments (BChls and carotenoids) are arranged in LH-II by using
VMD. It should take you about 45 minutes to complete.
LH-II from Rh. molischianum is a membrane protein
complex which absorbs light and transfers its energy to a reaction
center in a photosynthetic unit. It consists of eight copies of a
basic structural unit (building block) containing a heterodimer of
two protein subunits (![]() -apoprotein and
-apoprotein and ![]() -apoprotein),
three bacteriochlorophyls (BChls named B850a, B850b and B800,
according to their absorption wavelength in nm) and one
carotenoid. Eight of these units are assembled into the LH-II ring
as shown in Figure 1. In total, there are 16 protein
segments, 24 BChls (each contains a Mg
-apoprotein),
three bacteriochlorophyls (BChls named B850a, B850b and B800,
according to their absorption wavelength in nm) and one
carotenoid. Eight of these units are assembled into the LH-II ring
as shown in Figure 1. In total, there are 16 protein
segments, 24 BChls (each contains a Mg![]() ion at its center)
and 8 carotenoids.
ion at its center)
and 8 carotenoids.
You will start from viewing a building block of LH-II. From
there, you will then examine how two heterodimers fit together in
LH-II. Finally, you will investigate the complete LH-II ring which
is made of eight of these heterodimers.

In this subsection, you will examine the building block of
LH-II as shown in Figure 2 and investigate how the two
proteins and the four pigments are arranged.
![% latex2html id marker 2403
\framebox[\textwidth]{
\begin{minipage}{.2\textwidt...
...torial. You can start to answer questions in this subsection.}
\end{minipage} }](img14.gif)
In the pdb file of LH-II (lh2.pdb), there are eight
copies of the ![]() -apoprotein with segment names ALP1,
ALP2,.. .., ALP8 and eight copies of the
-apoprotein with segment names ALP1,
ALP2,.. .., ALP8 and eight copies of the ![]() -apoprotein with
segment names BET1, BET2,.. .., BET8. The corresponding
eight groups of BChls have segment names BCA1, BCA2,.. ..,
BCA8 and each consists of three individual BChls with residue
identification number (ResID) 57, 58 and 59
corresponding to BChls B850b, B800 and B850a respectively. Finally
there are eight carotenoids with segment names LYC1, LYC2,..
.., LYC8.
-apoprotein with
segment names BET1, BET2,.. .., BET8. The corresponding
eight groups of BChls have segment names BCA1, BCA2,.. ..,
BCA8 and each consists of three individual BChls with residue
identification number (ResID) 57, 58 and 59
corresponding to BChls B850b, B800 and B850a respectively. Finally
there are eight carotenoids with segment names LYC1, LYC2,..
.., LYC8.
You will first render a heterodimer of the two proteins
made of the segment ALP1 and BET1 in Cartoon
representation and color the ![]() - and
- and ![]() -apoproteins in
blue and purple respectively according to their segment names. You
will then render the four pigments contained in the heterodimer in
Licorice representation using various ways of coloring methods
which allow you to color the carotenoid in yellow and color BChl
B850a, B850b and B800 (resid 59,57 and 58) in red, green and pink,
respectively, to show their locations relative to each other.
Finally, you will render the two proteins in Surf
representation and color them so that you can learn how they pack
with each other and with the four pigments. Here is one way how to
do this all.
-apoproteins in
blue and purple respectively according to their segment names. You
will then render the four pigments contained in the heterodimer in
Licorice representation using various ways of coloring methods
which allow you to color the carotenoid in yellow and color BChl
B850a, B850b and B800 (resid 59,57 and 58) in red, green and pink,
respectively, to show their locations relative to each other.
Finally, you will render the two proteins in Surf
representation and color them so that you can learn how they pack
with each other and with the four pigments. Here is one way how to
do this all.
tbss> cd ![]() /tbss.work/photo-tutorial-files/1-excitation
/tbss.work/photo-tutorial-files/1-excitation
Open a VMD session and load the LH-II coordinate file lh2.pdb.
tbss> vmd lh2.pdb
Q1: What is the main secondary structure of
![]() - and
- and ![]() -apoproteins?
-apoproteins?
Q2: Where do ![]() - and
- and ![]() -apoproteins make
contact with each other?
-apoproteins make
contact with each other?
Q3: Can you tell that ![]() - or
- or
![]() -apoproteins are transmembrane proteins?
-apoproteins are transmembrane proteins?
Q4: What is the approximate distance between the
transmembrane helices of ![]() - and
- and ![]() -apoproteins?
-apoproteins?
Q5: Can you show that the N-terminals of both
proteins are on the same side of the membrane while their
C-terminals are on the other side.
You can see that ![]() - and
- and ![]() -apoproteins are
transmembrane proteins which consist mainly of
-apoproteins are
transmembrane proteins which consist mainly of ![]() -helices.
You also might have noticed that there is sufficient space in the
middle of the heterodimer to accommodate 3 BChls and 1 carotenoid
molecule. Now, let's have a look at how pigments are arranged in
the heterodimer.
-helices.
You also might have noticed that there is sufficient space in the
middle of the heterodimer to accommodate 3 BChls and 1 carotenoid
molecule. Now, let's have a look at how pigments are arranged in
the heterodimer.
NOTE: To delete the labels of atoms and distances on the
VMD OpenGL Display, Click Graphics in the VMD Main Window and select Labels. Select Atoms
to see a list of atom labels. Select atom names, then click Delete. Select Bonds to see a list of distance labels.
Select pairs of atom names, then click Delete.
Your LH-II structural unit should look like what is shown
in Figure 2 except that your BChls are rendered in their
full chemical structure.
Q6: What are the distances between (1) BChls B850a
and B850b; (2) BChls B850a and B800; (3) BChls B850b and B800?
Q7: What are the polar protein residues which make
close contact with the three BChls?
You can see that the BChls B850a (red), B850b (green) and
the carotenoid (yellow) are packed in the space between the
![]() - and
- and ![]() -apoproteins (blue and purple) while BChl
B800 make a contact to the N-terminal helix of the
-apoproteins (blue and purple) while BChl
B800 make a contact to the N-terminal helix of the
![]() -apoprotein. It can be seen that there are two histidine
side chains: one (
-apoprotein. It can be seen that there are two histidine
side chains: one (![]() His34) contacta BChl B850a and the other
(
His34) contacta BChl B850a and the other
(![]() His35) contacts BChl B850b. BChl 800 (pink) is coordinated
by an aspartate (
His35) contacts BChl B850b. BChl 800 (pink) is coordinated
by an aspartate (![]() Asp6). A very important is how two of
these building blocks assemble together. Experimentally, it has
been shown that these proteins actively self-assemble into LH-II.
Asp6). A very important is how two of
these building blocks assemble together. Experimentally, it has
been shown that these proteins actively self-assemble into LH-II.
In this section, you will study two LH-II structural units
from a VMD state prepared by us. You will then investigate the
packing of proteins and pigments between them.
Q1: How well do the two ![]() -apoproteins pack
with each other? How about the packing between the two
-apoproteins pack
with each other? How about the packing between the two
![]() -apoproteins?
-apoproteins?
Q2: What is the distance in Å (1) between two
![]() -apoproteins; (2) between two
-apoproteins; (2) between two ![]() -apoproteins?
-apoproteins?
Q3: What is the distance in Å between (1) two
BChls B850a (red); (2) two BChls B850b (green); (3) BChls B850a
(red, BCA1) and B850b'(green, BCA2) ; (4) B800 (pink, BCA1) and
B850b'; (5) B800 and B850a' (red, BCA2)?
It can be seen that the transmembrane helices of the
![]() -apoproteins are packed closely while the transmembrane
helices of the
-apoproteins are packed closely while the transmembrane
helices of the ![]() -apoproteins make very little contact with
each other. The termini of both proteins also make extensive
contact with each other. Can you imagine how a ring of eight of
these structural units looks like?
-apoproteins make very little contact with
each other. The termini of both proteins also make extensive
contact with each other. Can you imagine how a ring of eight of
these structural units looks like?
In this subsection, you will investigate an LH-II as a ring
of 8 heterodimers containing 24 BChls and 8 carotenoids.
Q1: What geometrical shape do the proteins and the
pigments in LH-II form? What are the advantages of having such an
arrangement?
Q2: What are the diameters of the rings of
![]() - and
- and ![]() -apoproteins in LH-II?
-apoproteins in LH-II?
Q3: Can you estimate the thickness of the membrane
(in Å) in which the LH-II is embedded?
Q4: What are the diameters of the B850 ring and the
B800 ring?
Q5: What are the distances between BChls B850a
(red) and its two adjacent BChls B850b (green)? Are they equal?
An LH-II is made up of eight heterodimers. They are
assembled into a circular ring with eight ![]() -apoproteins
(blue) forming an inner ring and eight
-apoproteins
(blue) forming an inner ring and eight ![]() -apoprotein (purple)
forming an outer ring. Pigments are embedded within the proteins
and are placed roughly in the middle of the transmembrane part of
the protein away from the bulk water at the interfaces of the
membrane. Sixteen BChls (B850a and B850b) and eight carotenoids
are packed between the rings of
-apoprotein (purple)
forming an outer ring. Pigments are embedded within the proteins
and are placed roughly in the middle of the transmembrane part of
the protein away from the bulk water at the interfaces of the
membrane. Sixteen BChls (B850a and B850b) and eight carotenoids
are packed between the rings of ![]() - and
- and ![]() -apoproteins
forming a ring of 16 BChls. Eight BChls B800 are held between
-apoproteins
forming a ring of 16 BChls. Eight BChls B800 are held between
![]() -apoproteins and supported by short helices from
-apoproteins and supported by short helices from
![]() -apoproteins. They form a larger ring of 8 BChls.
-apoproteins. They form a larger ring of 8 BChls.
In LH-II, light is absorbed by pigment molecules such as
BChls and carotenoids while proteins function as a scaffolding
structure that hold all the pigments in their places. In the next
section you will do some calculations to see how fast excitation
energy is transfered between pairs of BChls when their separation
is larger than their molecular sizes.
In this section, you are going to determine the
rate of excitation transfer between pairs of BChls within LH-II.
Your objective in this section is to learn how the rate of
excitation transfer between a pair of BChls is calculated. This
should take about half an hour to complete.
To do this, you will need a few mathematical formulae that
are used to calculate the excitation transfer rate. You will start
by determining the orientations of the transition dipole moments
of BChls in LH-II and represent them as cones using VMD. Then you
will use this structural information to compute the excitation
transfer rate between a pair of BChls using Mathematica.
Figure 4 shows the ring part of a BChl in LH-II.
Magnesium ions (Mg![]() ) are shown as white circles at the
center. The direction of the transition dipole moment is shown as
a cyan arrow. It points from the nitrogen atom of ring I (NB) to
that of ring III (ND).
) are shown as white circles at the
center. The direction of the transition dipole moment is shown as
a cyan arrow. It points from the nitrogen atom of ring I (NB) to
that of ring III (ND).
Figure 5 shows the top and side view of 24
BChls in LH-II forming two rings of BChls. The inner ring consists
of sixteen BChls (eight BChls B850a (red) and eight BChls B850b
(green) arranged alternatively). The outer ring consists of eight
BChls B800 colored in pink. Direction of the transition dipole
moments are represented by cones connecting two nitrogen atoms
NB and ND (see Figure 5). First, you
will learn how to compute an excitation transfer rate.
The rate ![]() of excitation energy transfer from BChl
i to BChl j can be determined via
of excitation energy transfer from BChl
i to BChl j can be determined via
where ![]() is the Coulomb coupling between the excited
state of BChl i and the ground state of BChl j;
is the Coulomb coupling between the excited
state of BChl i and the ground state of BChl j;
![]() is an overlap integral between the emission spectrum of
BChl i and the absorption spectrum of BChl j
which can be determined spectroscopically. For a pair of BChls
B850a
is an overlap integral between the emission spectrum of
BChl i and the absorption spectrum of BChl j
which can be determined spectroscopically. For a pair of BChls
B850a
![]() cm;
cm; ![]() = 3.1416 and
= 3.1416 and
![]() = 5.31 ps cm
= 5.31 ps cm![]() .
.
When the distance between the two BChls is larger than
their molecular sizes, the Coulomb interaction term ![]() can
be approximated as an induced dipole-induced dipole interaction
involving the transition dipole moments
can
be approximated as an induced dipole-induced dipole interaction
involving the transition dipole moments ![]() and
and
![]() of the two BChls whose central Mg
of the two BChls whose central Mg![]() ions are
separated by a distance
ions are
separated by a distance ![]() .
.
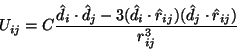
where ![]() ,
, ![]() and
and ![]() are unit
vectors along
are unit
vectors along ![]() ,
, ![]() and
and ![]() respectively; and C = 505,644 Å
respectively; and C = 505,644 Å![]() .cm
.cm![]() is a constant.
is a constant.
Let's ask a more specific question. What is the
excitation energy transfer rate between a pair of BChls B850a (say
segment names BCA1 and BCA2)? Let's call them the first and the
third BChls in the ring of 16 BChls. To be able to compute the
excitation energy transfer ![]() between the two BChls, we need
to determine the transition dipole moment
between the two BChls, we need
to determine the transition dipole moment ![]() and
and
![]() of the BChls and the relative position vector
of the BChls and the relative position vector
![]() . These vectors can be determined from the positions
of nitrogen atoms NB and ND and Mg
. These vectors can be determined from the positions
of nitrogen atoms NB and ND and Mg![]() ions (see
Figure 4).
ions (see
Figure 4).
In this subsection, you will use VMD to measure the
positions of the nitrogen atoms NB, ND and the
Mg![]() ions named MG of the BChls B850a from the segments BCA1 and BCA2 in the pdb file lh2.pdb. Their
positions will be used to determine the directions of the
transition dipole moments of the two BChls and the relative
position vectors between them. You will then draw the transition
dipole moments as cones to show how they are oriented in LH-II.
ions named MG of the BChls B850a from the segments BCA1 and BCA2 in the pdb file lh2.pdb. Their
positions will be used to determine the directions of the
transition dipole moments of the two BChls and the relative
position vectors between them. You will then draw the transition
dipole moments as cones to show how they are oriented in LH-II.
Q1: How is the transition dipole moment of a BChl
in the LH-II oriented relative to that of its neighboring
BChls.?
It can be seen that the BChls in LH-II have eight-fold
symmetry. The transition dipole moment of BChl B850a (red) points
roughly opposite to its neighboring BChl B850b (green) and it
points in the same direction to its adjacent BChl B800. Knowing
the positions of nitrogen atoms NB and ND and
Mg![]() ions, you can start to calculate the excitation transfer
rate in the next subsection.
ions, you can start to calculate the excitation transfer
rate in the next subsection.
In this final part, you will use the atomic positions
obtained from the previous subsection to calculate the excitation
transfer rate ![]() between two nearest BChls B850a (the first
and the third BChls in the ring of 16 BChls B850) using
Mathematica. You will also compute the excitation transfer rate
from BChl B800 to BChl B850a and the reverse transfer rate from
BChl B850a to BChl B800.
between two nearest BChls B850a (the first
and the third BChls in the ring of 16 BChls B850) using
Mathematica. You will also compute the excitation transfer rate
from BChl B800 to BChl B850a and the reverse transfer rate from
BChl B850a to BChl B800.
First, you need to calculate the directional vectors of the
BChl transition dipole moments ![]() ,
, ![]() and the
relative position vector
and the
relative position vector ![]() (both magnitude
(both magnitude ![]() and direction
and direction ![]() ) between the two BChls from segments
BCA1 and BCA2 in the LH-II coordinate file lh2.pdb. You will use these information to compute the induced
dipole-induced dipole interactions term
) between the two BChls from segments
BCA1 and BCA2 in the LH-II coordinate file lh2.pdb. You will use these information to compute the induced
dipole-induced dipole interactions term ![]() between them.
Then you will compute the transfer rate
between them.
Then you will compute the transfer rate ![]() between two BChls
B850a. Finally, you will determine the excitation transfer rate
between two BChls
B850a. Finally, you will determine the excitation transfer rate
![]() from BChl B800 to BChl B850a and the reverse transfer rate
from BChl B800 to BChl B850a and the reverse transfer rate
![]() .
.
tbss> cd ![]() /tbss.work/photo-tutorial-files
/tbss.work/photo-tutorial-files![]() 1-excitation
1-excitation
Open the mathematica file transfer.nb.
tbss> mathematica transfer.nb
This should open the mathematica file as a window shown in Figure 6.
STEP 1: Execute commands to define constants used
and load a Vector Analysis package. Click anywhere within the
cell covered by a blue bracket on the right which has a
semi-arrow. Then hold down the Shift key and press Enter. This will execute the command lines in the cell which
define some physical constants to be used in the calculation and
load a package called Vector Analysis into the program.
NOTE: Please ignore "Possible spelling error" message due
to the naming of variables used.
STEP 2: Calculate the directional vector
![]() of the transition dipole moment
of the transition dipole moment ![]() of the
BChl B850a in segment BCA1. In the Mathematica window, replace
the text {xNB1, yNB1, zNB1} and {xND1, yND1,
zND1} by the actual coordinates of nitrogen atoms NB and
ND of the BChl B850a in segment BCA1 read from the
previous subsection in the same format e.g. {1.23, 4.56, 7.89}.
Then execute the commands to calculate the directional vector
of the
BChl B850a in segment BCA1. In the Mathematica window, replace
the text {xNB1, yNB1, zNB1} and {xND1, yND1,
zND1} by the actual coordinates of nitrogen atoms NB and
ND of the BChl B850a in segment BCA1 read from the
previous subsection in the same format e.g. {1.23, 4.56, 7.89}.
Then execute the commands to calculate the directional vector
![]() by holding down the Shift key and then press Enter.
by holding down the Shift key and then press Enter.
STEP 3: Calculate the directional vector
![]() of the transition dipole moment
of the transition dipole moment ![]() of the
BChls B850a in segment BCA2. Repeat the previous step by
replacing the text {xNB3, yNB3, zNB3} and {xND3,
yND3, zND3} with the actual coordinates of nitrogen atoms NB and ND of the BChl B850a in segment BCA3 read
from the previous subsection in the same format e.g. {1.23, 4.56,
7.89}. Then execute the commands to calculate the directional
vector
of the
BChls B850a in segment BCA2. Repeat the previous step by
replacing the text {xNB3, yNB3, zNB3} and {xND3,
yND3, zND3} with the actual coordinates of nitrogen atoms NB and ND of the BChl B850a in segment BCA3 read
from the previous subsection in the same format e.g. {1.23, 4.56,
7.89}. Then execute the commands to calculate the directional
vector
![]() by holding down the
Shift key and then press Enter.
by holding down the
Shift key and then press Enter.
STEP 4: Compute the separation vector
![]() and the distance
and the distance ![]() (Å) between the the
central Mg
(Å) between the the
central Mg![]() ions of the two BChls B850a. (NOTE: In the
mathematica script, the distance
ions of the two BChls B850a. (NOTE: In the
mathematica script, the distance ![]() is denoted as L13).
Replace the text {xMG1, yMG1, zMG1} and {xMG3,
yMG3, zMG3} by the actual coordinates of the two Mg
is denoted as L13).
Replace the text {xMG1, yMG1, zMG1} and {xMG3,
yMG3, zMG3} by the actual coordinates of the two Mg![]() ions, named MG , on the two BChls B850a read from the
previous subsection in the same format e.g. {1.23, 4.56, 7.89}.
Then execute the commands to calculate the relative position
vector of
ions, named MG , on the two BChls B850a read from the
previous subsection in the same format e.g. {1.23, 4.56, 7.89}.
Then execute the commands to calculate the relative position
vector of
![]() and the
separation distance
and the
separation distance
![]() (measured in Å) by holding down the Shift key and then
press Enter.
(measured in Å) by holding down the Shift key and then
press Enter.
Q1: What is the separation distance between two
BChls B850a? How large is it compare with the size of a BChl? Why
is this important?
STEP 5: Calculate the directional vector
![]() of the relative position vector
of the relative position vector ![]() .
Execute the commands to calculate the directional vector
.
Execute the commands to calculate the directional vector
![]() of the relative position vector
of the relative position vector ![]() i.e.
i.e.
![]() by holding down
the Shift key and then press Enter.
by holding down
the Shift key and then press Enter.
STEP 6: Calculate the induced dipole-induced dipole
interaction term ![]() . (cm
. (cm![]() ) Execute the commands to
compute the induced dipole-induced dipole interaction term
) Execute the commands to
compute the induced dipole-induced dipole interaction term
![]() calculated according to the second equation above by
holding down the Shift key and then press Enter.
calculated according to the second equation above by
holding down the Shift key and then press Enter.
STEP 7: Calculate the excitation transfer rate
![]() (per ps). Execute this section to compute the the
excitation transfer rate
(per ps). Execute this section to compute the the
excitation transfer rate ![]() according to the first equation
above by holding down the Shift key and then press Enter.
according to the first equation
above by holding down the Shift key and then press Enter.
Q2: What is the rate of excitation transfer between
two BChls B850a?
Now let's calculate the excitation transfer rate ![]() from
BChl B800 to Bchl B850a and the reverse rate
from
BChl B800 to Bchl B850a and the reverse rate ![]() .
.
STEP 8: Calculate the forward rate ![]() of
excitation transfer from a BChl B800 to its nearest BChl B850a.
Up to here we have determined all the quantities required for the
calculation of the transfer rate from the BChl B800 of segment
BCA1 to its nearest BChl B850a of the same segment for you.
You just need to execute the commands to calculate the rate of
forward energy transfer by holding down the Shift key and
then press Enter.
of
excitation transfer from a BChl B800 to its nearest BChl B850a.
Up to here we have determined all the quantities required for the
calculation of the transfer rate from the BChl B800 of segment
BCA1 to its nearest BChl B850a of the same segment for you.
You just need to execute the commands to calculate the rate of
forward energy transfer by holding down the Shift key and
then press Enter.
STEP 9: Calculate the rate ![]() of excitation
transfer from a BChl B850a to BChl B800. Similarly in this step,
we have determined all the quantities required for the calculation
of the transfer rate from the BChl B850a of segment BCA1 to
its nearest BChl B800 of the same segment for you. You just need
to execute the commands to calculate the backward transfer rate by
holding down the Shift key and then press Enter.
of excitation
transfer from a BChl B850a to BChl B800. Similarly in this step,
we have determined all the quantities required for the calculation
of the transfer rate from the BChl B850a of segment BCA1 to
its nearest BChl B800 of the same segment for you. You just need
to execute the commands to calculate the backward transfer rate by
holding down the Shift key and then press Enter.
Q3: Compare the forward excitation transfer rate
![]() with the reverse transfer rate
with the reverse transfer rate ![]() . Which direction of
excitation transfer is more favorable?
. Which direction of
excitation transfer is more favorable?
You can see that the excitation transfer rate from BChl
B800 to BChl B850a is six orders of magnitude higher than that of
the reverse direction. So it is much more favorable for excitation
energy to be transferred from a ring of 8 BChls B800 to a ring of
16 BChls B850a within an LH-II. The factor that directly affects
the forward and reverse rates is the overlap integral ![]() which depends on the energies of the emission peak
which depends on the energies of the emission peak ![]() and
absorption peak
and
absorption peak ![]() . When
. When ![]() is more than
is more than ![]() you get large
value of overlap integral and vice versa.
you get large
value of overlap integral and vice versa.