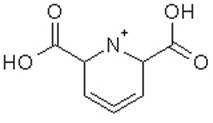
From: Chris Harrison (charris5_at_nd.edu)
Date: Wed Jan 14 2009 - 23:25:19 CST
This is an organic compound. Is the image missing some atoms .... the
counter-ion?
If the parameters are available, then NAMD can simulate the molecule.
Please look in the CHARMM forcefield to see if this is a possibility.
Alternatively, you can parameterize the molecule yourself. The paratool
plugin in VMD can be of help with this; however you will still need to
understand how to do the process manually. Please look at:
1. MacKerell, et al; J Phys Chem B 102:3586 (1998)
2. MacKerell, Wiorkiewicz-Kuczera, & Karplus; JACS 117:11946 (1995)
C.
-- Chris Harrison, Ph.D. Theoretical and Computational Biophysics Group NIH Resource for Macromolecular Modeling and Bioinformatics Beckman Institute for Advanced Science and Technology University of Illinois, 405 N. Mathews Ave., Urbana, IL 61801 char_at_ks.uiuc.edu Voice: 217-244-1733 http://www.ks.uiuc.edu/~char Fax: 217-244-6078 On Thu, Jan 8, 2009 at 12:20 PM, Casey,Richard <Richard.Casey_at_colostate.edu>wrote: > Hello, > > > > We would like to run molecular dynamics simulations for the inorganic > compound shown below (and attached) using NAMD/VMD. Can inorganic compounds > be modeled with NAMD? If so, what would be an appropriate force field > (Charmm? Amber? Other?) > > > > > > > > > > -------------------------------------------- > > Richard > > >

This archive was generated by hypermail 2.1.6 : Wed Feb 29 2012 - 15:52:15 CST