Collaborations between structural biology researchers, in general, focus on specific biopolymer systems, e.g., on a particular class of proteins. In the course of such investigations, researchers collect information as they carry out experiments, run simulations, perform analyses, and discuss results. Accordingly, an intuitive paradigm for collaboratory software is the project. Hence, BioCoRE represents the information contributed by collaborators as a project, and makes projects the fundamental units of organization. Each project contains all the documents, results, history, and communications for a specified set of collaborating scientists.
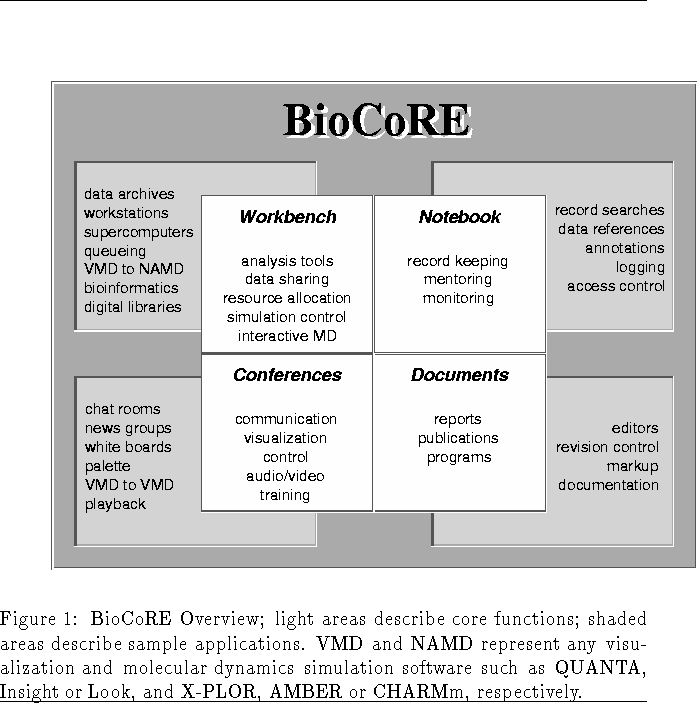
BioCoRE will be accessible via a Java-enabled web browser. The user will log into the system via a browser and will be presented with a menu containing all projects for which access is authorized. The user will then select a project, which is created and stored in a BioCoRE collaboratory server at a selected web site. At this point the user has access to the main BioCoRE user environment, summarized in Figure 1, which supports four main types of user activities:
The common user interface will be able to launch the Workbench, Notebook, Conferences, and Documents interfaces for the selected project. The functionality of these interfaces, as will be implemented in BioCoRE, is described below.
A key portion of any computational structural biology project is the use of simulation, analysis, and visualization software. Quite often it is desirable to access these programs during a collaborative session, to perform such tasks as analysis of existing data, investigations with new data or parameters, or visualization of results. The Workbench interface will give the user access to modeling software tools, such as the simulation or visualization programs of preference, and will support data sharing, resource allocation, simulation control, and interactive simulation. The collaboratory Workbench will provide the user with transparent access to all collaboratory data and computational tools without regard to where data are stored or where programs run while automating much of the log-keeping required for simulations.
As an example, consider two researchers working together online who
wish to access the results of a protein simulation currently running
with the Workbench control environment. Using the
Workbench controls, both researchers can start separate
visualization applications that will connect to a running simulation
and display the same data; the two researchers may actually utilize
different molecular graphics programs, e.g. VMD and Quanta ( Polygen, 1988
).
( Polygen, 1988
).
Many different structural biology simulation methods, such as molecular dynamics simulations, generate large amounts of data to be managed and stored. Large files will be kept in the collaboratory archives that will store all the project data available in the system (aside from Notebook entries) and will be presented as an unified directory structure. Data added to the archive will be logged in the Notebook, together with comments. These Notebook entries will allow searching of all collaboratory data by topic without burdening the Notebook with large files. Data in the archive will be accessed by either manual download or automatically by collaboratory-enabled software or collaboratory software wrappers.
Third-party interactive software packages are an important tool in the modern scientific process and their use will be simplified by the collaboratory environment. Interactive software launched from within the environment will be wrapped to allow access to data in the archive which will be automatically downloaded into a special working directory, while the results will be automatically archived and logged.
BioCoRE will simplify running jobs on remote resources, as part of the Workbench capabilities. After selecting an available tool, such as a simulation program, the user will specify input and output files in the archive and enter configuration information. The Workbench will then present the user a choice of suitable resources for fulfilling the request such as the user's workstation, a remote workstation, or a dedicated compute server. The job will be executed as directed by the user and logged in the Notebook along with comments and accounting information.
The Workbench will also provide interactive steering interfaces. A primary example of this is the interface between VMD and NAMD. Selecting from a list of running jobs, the user will be presented with an appropriate monitoring and manipulation interface for the selected job. Any modifications to the progress of a simulation will be recorded in the Notebook, ensuring that all inputs to the simulation are saved.
Some specific tools to be incorporated into the Workbench include VMD, NAMD, MDTools , and BLAST and FASTA searches, or interfaces to SWISS-PROT ( Appel et al., 1994 ), PIR ( Sidman et al., 1988 ), and PDB ( Bernstein et al., 1977 ), available through the Biology Workbench.
The Notebook interface will provide the record keeping, mentoring, and monitoring capabilities of the collaboratory. The interactive collaboratory Notebook will present the user with a chronological view of the entire project. The Notebook will be organized into entries. A Notebook entry will comprise a series of references to components. Any form of data recognized by the collaboratory environment will be accepted as a component, that is handled by the Notebook according to its type. For example, rather than pasting a molecular coordinate file or an image in the Notebook, a visualization program configuration is included which will launch an appropriate viewer with a preselected representation and orientation. Component types recognized by the collaboratory environment will include text or HTML documents, images, tables of data displayed as interactive graphs, visualization program configurations displayed as a thumbnail, simulation trajectories displayed as a summary, and lists of data references to other entries or their components.
It will be possible to search for entries in the Notebook by title, author, date, subproject, topic, and type as well as component properties such as content, or entries referred to. The user will also be presented with ready access to several standard reference lists such as calendar views by day, week, and month, unread and recently visited entries, and a palette of references chosen by the user.
The Conferences interface will meet requirements for communication, visualization control, audio/video capabilities, and training. This interface will allow a user to review ongoing Conferences which this user is eligible to join, or to create a new conference and invite others to join it. Upon joining a conference, the user will be presented with a menu of third-party conference tools which can be launched from within the environment. These tools will automatically connect the user to other conference participants. The user will also be presented with the conference palette which will contain references to Notebook entries being discussed and to the Notebook entry containing the ongoing conference log. Users may dynamically enter and leave a conference, and a conference may continue asynchronously through posting of messages and replies.
A major conferencing tool to be developed will be the ability to slave one VMD session to another such that their displays are identical. This will enable one collaborator to control visualization on other collaborators' displays during a conference. The control of the visualization can be passed from one collaborator to another.
The Documents interface will meet requirements for preparing reports, publications, and other forms of dissemination. These features can also aid in software development. The key requirement is multi-author revision control services. This will most likely be accomplished by wrapping the existing CVS (Concurrent Versions System) package in such a way that existing tools with CVS interfaces can be used without modification. Added value will be provided by incorporating CVS logs into the Notebook. Additional features will distribute and archive commentary and markups of drafts.