


Next: Structural Alignment of Aquaporins
Up: Aquaporin Tutorial
Previous: Introduction
Contents
Subsections
Introduction to Aquaporin Structure
In this unit you will use the conventional molecular graphics tools of VMD to
become familiar with the key structural features of aquaporins,
and how these are related to aquaporin function.
You will first consider bovine aquaporin (PDB code 1J4N).
In order to learn about the structural features of aquaporins, you will create
several graphical representations of the molecule. Here, we will only guide you
through the steps to create the necessary representations. If you want to know
more about graphical representations, please look at the main VMD
tutorial3.
- 1
- Choose the File
 New Molecule... menu item in the
VMD Main window. Another window, the Molecule File Browser,
will appear on your screen.
New Molecule... menu item in the
VMD Main window. Another window, the Molecule File Browser,
will appear on your screen.
- 2
- Use the Browse... button to find the file 1j4n.pdb in
the directory
aqp-tutorial-files  PDB in
the tutorial directory. Note that when you select the file, you will be back
in the Molecule File Browser window. In order to actually load the
file you have to press Load. Do not forget to do this!
PDB in
the tutorial directory. Note that when you select the file, you will be back
in the Molecule File Browser window. In order to actually load the
file you have to press Load. Do not forget to do this!
Now, the aquaporin is shown on your screen in the OpenGL Display
window. You may close the Molecule File Browser window at any time.
- 3
- Choose the Graphics
 Representations... menu
item. A window called Graphical Representations will appear and
you will see the current graphical representation used to display your molecule
highlighted in light green.
Representations... menu
item. A window called Graphical Representations will appear and
you will see the current graphical representation used to display your molecule
highlighted in light green.
- 4
- In the Draw Style tab you can change the style
and color of the representation. In the Selected Atoms text entry
of the Graphical Representations window, delete the word all.
In the place of all, type protein and press the Apply button
in the bottom right-hand corner of the window, or hit Enter or Return key on your
keyboard. It is important that you do this every time you type something in Selected Atoms.
From Drawing Method,
choose the Tube menu item. The representation draws a tube along
the backbone of the protein (Fig. 1). Now, change the color of
the molecule by choosing the Molecule menu item from the Coloring Method menu.
Figure:
Tube representation of aquaporin structure.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2 in]{pictures/reptube}
}
\end{center}\vspace{-0.7cm}\end{figure}](img9.gif) |
The tube representation you created will be used throughout the tutorial to
look at the structural alignment of aquaporins, but before you start using
the Multiseq program, you will create other representations in order to learn some of the
important structural features of aquaporins.
- 5
- In the Graphical Representations window, click the Create Rep button. This will create a new representation, identical to the
one you had before. This time, choose the drawing method NewCartoon. This drawing method shows the secondary structure of aquaporins
(Figure 2).
Figure:
The NewCartoon representation of an aquaporin structure.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2 in]{pictures/repcartoon}
}
\end{center}\vspace{-0.7cm}\end{figure}](img10.gif) |
- 6
- In the Graphical Representations window, double-click on the
first representation. This will make the text of the first representation red in this menu,
and will hide the corresponding representation in the OpenGL Display window.
- 7
- Click on the OpenGL window and use your mouse to rotate the molecule and look at the structure of aquaporins.
For the rest of this section, you will continue to look at the aquaporin
structure in Tube representation. You can always go back and look at the
NewCartoon representation, if necessary.
- 8
- In the Graphical Representations window, double click on the
first representation. This will show it again in the OpenGL window. Double
click on the second representation (NewCartoon) to hide it.
Now look closer at the structural details of the aquaporin structural elements.
- 9
- Create a new representation, clicking on the Create Rep button. This time, you will focus
on the helices of the reentrant loops mentioned above (see box). In the Selected Atoms text entry of the
Graphical Representations window type resid 79 to 88 196 to 204. These residues correspond to two helices of
aquaporin that face each other in the middle of the channel. Choose the coloring method ColorID
 1 and the drawing method Tube.
1 and the drawing method Tube.
- 10
- Repeat step 9. This time in Selected Atoms, type resid 74 to 78 191 to 195, and for the Coloring Method, select ColorID
 7. These residues correspond to the extended polypeptide regions of the
reentrant loops of aquaporins (see box).
7. These residues correspond to the extended polypeptide regions of the
reentrant loops of aquaporins (see box).
Now that you have localized the reentrant loops, the rest of the protein will appear dim.
- 11
- Click (once) on the first representation. In the Material menu,
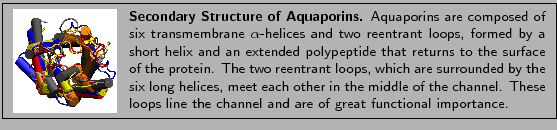
choose Transparent. Your OpenGL window should look like Fig. 3.
Figure:
The reentrant loops are a key structural feature of
aquaporins. Each reentrant loop is formed by a short helix, shown in red, and
an extended polypeptide, shown in green.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2 in]{pictures/repreent}
}
\end{center}\vspace{-0.7cm}\end{figure}](img12.gif) |
Now that you can locate the reentrant loops, you will examine the NPA motif within
the reentrant loops.
- 12
- Create a new representation, by clicking on the Create Rep button. Make sure the new representation is not transparent.
In the Selected Atoms text entry
of the Graphical Representations window, type resid 78 to 80 194
to 196. These residues correspond to the two NPA motifs present in
aquaporin. Choose the drawing method Licorice and coloring method
Type. Look at the NPA motifs, can you see how they would be
hydrogen bonded?
Figure:
The NPA motifs in aquaporins.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.8 in]{pictures/repnpa}
}
\end{center}\vspace{-0.7cm}\end{figure}](img14.gif) |
- 13
- Create another representation by clicking the Create Rep button. With
the same coloring and drawing methods of the previous selection, type resid 197 in the Selected Atoms text window. This will draw an
important arginine residue in aquaporins.
Figure:
The conserved arginine provides H-bond sites to water molecules inside the channel.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.8 in]{pictures/reparginine}
}
\end{center}\vspace{-0.7cm}\end{figure}](img15.gif) |
- 14
- Repeat step 13, but this time type resname GLU in the
Selected Atoms text window. This will select all the glutamates in
the protein. Notice that there are only two glutamate side chains in the
transmembrane region that are in the helical core of the protein. If you look
closely (c.f. Fig. 6) you will see that each one of these
glutamates sits behind a reentrant loop.
Figure:
The glutamates help stabilize the reentrant loops.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2 in]{pictures/repglu}
}
\end{center}\vspace{-0.7cm}\end{figure}](img18.gif) |
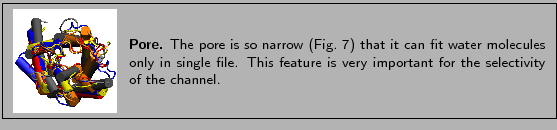
Finally, you are going to look into the aquaporin pore.
- 15
- Create a representation. In the Selected Atoms text window
type protein. Set the drawing method to VDW
(van der Waals). Each atom is now represented by a sphere. In this
way you can view the volumetric distribution of the protein.
- 16
- Rotate the molecule so that you can see it from the top. Look down the
pore. Can you see the pore? How many water molecules you think fit there?
Figure:
The pore of aquaporins can only accomodate water molecules in single file.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2 in]{pictures/repvdw}
}
\end{center}\vspace{-0.7cm}\end{figure}](img19.gif) |
- 17
- Create a new representation. In the Selected Atoms text window
type water. Set the Drawing Method to VDW and the Coloring
Method to type. This will create a representation with all the water
molecules the crystallographers resolved in the crystal structure. You can now
see that the pore is filled with water molecules forming a single file.
Now that you have learned about the structural features of aquaporins, you
will load the other three aquaporins mentioned above and get ready to align them. Before that,
you should turn off all representations created so far, leaving only the
first.
- 18
- In the Graphical Representations window, double click on each of
the graphical representations, except the first. This will make them
appear in light color in this menu, and will hide the corresponding views in the
OpenGL window. Select the first representation and set the material to
Opaque.
In this tutorial you will be comparing four aquaporin structures listed in Table 1.2. You have already loaded the 1j4n molecule. Now, you will load
the other three aquaporins.
Table 1:
Aquaporin structures
| PDB code |
Description |
| 1j4n |
Bovine AQP1 |
| 1fqy |
Human AQP1 |
| 1lda |
E. coli Glycerol Facilitator (GlpF) |
| 1rc2 |
E. coli AqpZ |
|
- 1
- The remaining aquaporins, 1fqy, 1lda,
1rc2, need to be loaded into VMD and have their molecule's graphical representations individually changed to
Tube. To do this, you need to refer back to the previous section,
Structure and Function of Aquaporins, and repeat steps 1 through
4. Make sure that each PDB is loaded into a new molecule.
Figure:
Aquaporin structures loaded into VMD.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=3 in]{pictures/loaded}
}
\end{center}\vspace{-0.7cm}\end{figure}](img21.gif) |
You should now see the four molecules loaded in the
OpenGL Display (Figure 8). If you do not see all molecules, hit S on the keyboard while in the OpenGL display. Then use your mouse to scale the molecules in the display, such that you can see all four. Note that the structures are not aligned, but
rather placed according to the coordinates specified in their PDB files.
- 2
- Close the Graphical Representations window and Molecule File Browser window.
You are now ready to start performing Multiple Sequence Alignment.



Next: Structural Alignment of Aquaporins
Up: Aquaporin Tutorial
Previous: Introduction
Contents
school@ks.uiuc.edu
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...on (see VMD tutorial) and resume your
study at a later time.}
\end{minipage} }](img6.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...on (see VMD tutorial) and resume your
study at a later time.}
\end{minipage} }](img6.gif)
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2 in]{pictures/repreent}
}
\end{center}\vspace{-0.7cm}\end{figure}](img12.gif)



![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...lude only one copy of the molecule in the
provided pdb file.}
\end{minipage} }](img22.gif)