


Next: Phylogenetic Tree
Up: AARS Tutorial
Previous: Comparing Protein Structure And
Contents
Subsections
Starting up Residue Selection
The Residue Selection feature lets you analyze conservation,
using different measures, and highlight residues in the Sequence Display and Structure Display simultaneously.
Residue Selection allows you to examine the conservation on
a per residue basis.
Figure 11:
To access Residue Selection:
|
- 1
- Click on Tools menu in the main Multiple Alignment window top pull-down menu.
- 2
- Select Residue Selection.
|
From this pull-down menu item you can select Q Value or Sequence Identity.
Each tool has its own window that allows you to select a a greater than or less than value between 0.0 to 1.0.
For this exercise, we will first examine structure conservation. Make sure Molecule Coloring is set to Q per residue.
- 1
- In the main Multiple Alignment window top pull-down, select Tools
 Residue
Selection Residue
Selection  Q Value. Q Value.
- 2
- A new window entitled Q Value will appear. Select Greater than
- 3
- To the right, you can select a value between 0.0 to 1.0. Select 0.5 for a value.
- 4
- Then, click on the Select button.
|
Look in the OpenGL Display window and see what happens to the molecule. Since you selected the Q Value to be greater than or equal to 0.5, the majority of the molecule
will be yellow. Also, the Sequence Display, in the Multiple Alignment window, will have the majority of
the molecules
highlighted. Why is this? By selecting the Q Value to be greater than or equal to 0.5, you
have selected structural conservation at Q=0.5 or greater. In other words, you have
demonstrated where the Q Value, structural conservation, is at a moderate to high level
in all 4 aligned molecules - almost obscuring where high levels of conservation occur.
Due to the high level of conservation, the Bond highlight style is not very informative.
Figure 13:
Q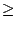 0.5 with Bonds Highlight Style
0.5 with Bonds Highlight Style
|
|
You can change this by accessing the main Multiple Alignment window top pull-down menu:
- 5
- Clicking on View
 Highlight Style.
Highlight Style.
- 6
- A side menu will appear alongside Highlight Style. Select Trace for the highlight style.
Figure 14:
Q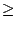 0.5 with Trace Highlight Style
0.5 with Trace Highlight Style
|
|
As you can see, the areas with high levels of conservation are easily discerned using the Trace style. Now go back to the Q Value window and click on the Close Window button.
Before beginning this exercise, change the highlight style back to Bonds and the Molecule Coloring to Sequence Identity per residue.
In order to display sequence conservation, follow these steps.
- 1
- Go back to the main Multiple Alignment window and select Tools
 Residue Selection. Residue Selection.
- 2
- Select Sequence Identity.
- 3
- In the Sequence Identity window, set the measure to Greater than.
- 4
- Select the value 0.5.
- 5
- Click the Select button.
|
Do you notice a difference in the highlighted areas, in comparing the structure (Q) to
sequence? Upon examining the residues in the Sequence display, notice how the majority of
the sequence conservation occurs among the top 3 molecules. Since the top 3 molecules are
AspRS and the bottom is SerRS, this pattern of sequence conservation makes sense.
Go back to the Sequence Identity window. Decrease the value to
0.2 and click Select. Take note of how many conserved residues are in the OpenGL Display and Sequence Display.
Now, increase the value back to 0.5. You will see, as you progress, the yellow color diminishing
among the molecules.
Figure 16:
Sequence Identity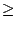 0.7
0.7
|
|
The yellow coloring remains towards the core of the molecule and at the dimerization site.
Increase the value to 0.7 and click Select; you should notice that the only highlighted sequences in the Sequence Display are those that match exactly.
Finally, change the value to 1.0. Between Sequence Identity 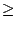 0.7 and Sequence Identity
0.7 and Sequence Identity 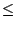 1.0, there are only 10 residues which are
identical and span all four molecules. Go back to the Sequence Identity window and click on the Close Window button.
1.0, there are only 10 residues which are
identical and span all four molecules. Go back to the Sequence Identity window and click on the Close Window button.
Figure 17:
Highlighted residues in Sequence Display
|
|
This exercise will further examine structure conservation. Make sure to change Molecule Coloring
back to Q per residue.
- 1
- Go back to the main Multiple Alignment window top pull-down menu and select Tools
 Residue Selection. Residue Selection.
- 2
- Select Q Value.
- 3
- Select Less than and 0.1 value.
- 4
- Then click on the Select button.
|
If you had set Molecule Coloring to Q per residue you will notice that the majority
of the red areas have turned yellow, and the blue is visible. If you look in the Sequence Display, you
will see that the majority of SerRS residues have been highlighted. By using these settings in Q
value, you have demonstrated where the aligned molecules have the least structural
conservation,Q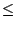 0.1.
0.1.
Figure 19:
Q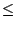 0.1
0.1
|
|
Now select Greater than or equal to and 0.6 in the Q Value window , and click on Select. Notice, in
the OpenGL Display that the majority of the blue area inside the molecules has turned yellow. Also,
note what has occurred in the sequence display. Many areas of the AspRS
molecules are highlighted. Occasionally, an amino acid in SerRS is
highlighted along with those of the AspRS. As you increase the value to 0.7, there are no
highlighted residues from SerRS. Now that we have completed the exercises in Residue Selection, close the
Q Value window.
How does this analysis indicate which molecules are more closely related to each other?
In Exercise 1, we evaluated structure conservation on a moderate to high level and noticed
when we changed highlight style that the structural conservation is highest in the core. Sequence
conservation, in Exercise 2, was also highest in the core of the molecule and at the dimerization
site. We then further analyzed structure conservation during Exercise 3. We noticed that upon
increasing the value of structure conservation, the highest structural conservation is among the three AspRSs. When the Q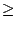 0.7,
no residues is SerRS had Q
0.7,
no residues is SerRS had Q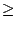 0.7 with the three AspRS, indicating poor structure conservation. The three AspRS molecules
are more closely related to each other in comparison to SerRS.
0.7 with the three AspRS, indicating poor structure conservation. The three AspRS molecules
are more closely related to each other in comparison to SerRS.
Having examined the relationship between the four AARSs using structure and sequence conservation,
we will now use phylogenetic trees to display the evolutionary relationships graphically.



Next: Phylogenetic Tree
Up: AARS Tutorial
Previous: Comparing Protein Structure And
Contents
workshop+urbana@ks.uiuc.edu