

Next: Trajectories and Movie Making
Up: VMD Tutorial
Previous: Introduction
Subsections
Working with a Single Molecule
In this section you will learn the basic functions of VMD. We will start with loading a molecule, displaying the molecule, and rendering publication-quality molecule images. This section uses the protein ubiquitin as an example molecule. Ubiquitin is a small protein responsible for labeling proteins for degradation, and is found in all eukaryotes with nearly identical sequences and structures.
Loading a Molecule
The first step is to load our molecule. A pdb
file,
1ubq.pdb (Vijay-Kumar et al., JMB, 194:531, 1987),
that contains the atom coordinates of ubiquitin is
provided with the tutorial.
Figure 4:
Loading a Molecule.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_MoleFileBrow}
}
\end{center}
\end{figure}](img9.gif) |
- 1
- Start a VMD session. In the VMD Main window, choose File
 New Molecule... (Fig. 4(a)). Another window, the Molecule
File Browser window (Fig. 4(b)), will appear on your screen.
New Molecule... (Fig. 4(a)). Another window, the Molecule
File Browser window (Fig. 4(b)), will appear on your screen.
- 2
- Use the Browse... (Fig. 4(c))
button to find the file
1ubq.pdb in vmd-tutorial-files directory. Note that when you select
the file, you will be back in the Molecule File Browser window.
In order to actually load the file you have to press Load
(Fig. 4(d)). Do not forget to do this!
Now, ubiquitin is shown in the OpenGL Display window. You may close the Molecule File Browser window
at any time.
Displaying the Molecule
In order to see the 3D structure of our protein, we will use the mouse
in multiple modes to change the viewpoint.
VMD allows users to rotate, scale and translate
the viewpoint of your molecule.
Figure 5:
Rotation modes. (a) Rotation axes when holding down the left mouse key. (b) The rotation axis when holding down the right mouse key.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_rotations}
}
\end{center}
\end{figure}](img12.gif) |
- 1
- In the OpenGL Display, press the left mouse button down and move the mouse. Explore what
happens. This is the rotation mode of the mouse and allows you to rotate
the molecule around an axis parallel to the screen (Fig.
5(a)).
- 2
- If you hold down the right mouse button and repeat the previous step,
the rotation will be done around an axis perpendicular to your screen (Fig.
5(b)) (For Mac users, the right mouse button
is equivalent to holding down the command key while pressing the mouse button).
- 3
- In the VMD Main window, look at the Mouse menu (Fig. 6). Here, you
will be able to switch the mouse mode from Rotation
to Translation or Scale modes.
Figure 6:
Mouse modes and their characteristic cursors.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_mouse}
}
\end{center}
\end{figure}](img14.gif) |
- 4
- Choose the Translation
mode and go back to the OpenGL Display. You can now move the molecule around when you
hold the left mouse button down.
- 5
- Go back to the the Mouse menu and choose the Scale
mode this time. This will allow you to zoom in or out by moving the mouse horizontally
while holding the left mouse button down.
It should be noted that these actions performed with the mouse
only change your viewpoint and do not change any of the atomic coordinates
associated with the molecule.
Another useful option is the Mouse
 Center menu item. It allows you to specify the point around which rotations are done.
Center menu item. It allows you to specify the point around which rotations are done.
- 6
- Select the Center menu item and pick one atom at one of the
ends of the protein; The cursor should display a cross.
- 7
- Now, press r, rotate the molecule with the mouse and see how
your molecule moves around the point you have selected.
- 8
- In the VMD Main window, select the Display
 Reset View menu item to
return to the default view. You can also reset the view by pressing the ``=" key when you are in the OpenGL Display window.
Reset View menu item to
return to the default view. You can also reset the view by pressing the ``=" key when you are in the OpenGL Display window.
VMD can display your molecule in various ways by the Graphical Representations shown in Fig. 7. Each representation is defined by four main parameters: the selection of atoms included in the representation, the drawing style, the coloring method, and the material. The selection determines which part of the molecule is drawn, the drawing method defines which graphical representation is used, the coloring method gives the the color of each part of the representation, and the material determines the effects of lighting, shading, and transparency on the representation. Let's first explore different drawing styles.
- 1
- In the VMD Main window, choose the Graphics
 Representations... menu
item. A window called Graphical Representations will appear and
you will see highlighted in yellow (Fig. 7(a)) the current default representation displaying your molecule.
Representations... menu
item. A window called Graphical Representations will appear and
you will see highlighted in yellow (Fig. 7(a)) the current default representation displaying your molecule.
- 2
- In the Draw Style tab (Fig. 7(b)) we can
change the style (Fig. 7(d))
and color (Fig. 7(c)) of the representation. In this section we will focus in
the drawing style (the default is Lines).
- 3
- Each Drawing Method has its own parameters. For instance, change
the Thickness of the lines by using the controls
on the lower right-hand-side corner (Fig. 7(c)) of the
Graphical Representations window.
Figure 7:
The Graphical Representations window.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_GrapRepr}
}
\end{center}
\end{figure}](img16.gif) |
- 4
- Click on the Drawing Method (Fig. 7(d)), and you will see a list of options. Choose VDW (van der Waals). Each atom is now represented by a sphere,
allowing you to see more easily the volumetric distribution of the protein.
- 5
- When you choose VDW for drawing method, two new controls would show up in the lower right-hand-side corner (Fig. 7(e)). Use these controls to change the
Sphere Scale to 0.5
and the Sphere Resolution to 13. Be aware that the higher
the resolution, the slower the display of your molecule will be.
- 6
- Press the Default
button. This allows you to return to the default
properties of the chosen drawing method.
The previous representations allow you to see the micromolecular
details of your protein by displaying every single atom. More general structural
properties can be demonstrated better by using more abstract drawing methods.
- 7
- Choose the Tube style under Drawing Method and observe the backbone of your
protein. Set the Radius at 0.8. You should get something similar to Fig. 8.
- 8
- By looking at your protein in the tube drawing method, see if you can distinguish the helices,
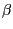 -sheets and coils present in the protein.
-sheets and coils present in the protein.
Figure 8:
Licorice (left), Tube (center) and NewCartoon (right) representations of Ubiquitin
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=0.8\textwidth]{FIGS/u2_LicoTubeNewC}
}
\end{center}
\end{figure}](img18.gif) |
The last drawing method we will explore is NewCartoon. It gives a simplified representation of a protein based in its
secondary structure. Helices are drawn as coiled ribbons, 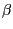 -sheets as solid
arrows and all other structures as a tube. This is probably the most popular
drawing method to view the overall architecture of a protein.
-sheets as solid
arrows and all other structures as a tube. This is probably the most popular
drawing method to view the overall architecture of a protein.
- 9
- In the Graphical Representations window, choose Drawing Method
 NewCartoon. You can now easily identify how many helices,
NewCartoon. You can now easily identify how many helices, 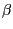 -sheets and coils are present in
the protein.
-sheets and coils are present in
the protein.
Now, let's explore different coloring methods for our representations.
- 10
- In the Graphical Representations window, you can see that the default coloring method is Coloring Method
 Name. In this coloring method, if you choose a drawing method that shows individual atoms, you can see that they have different colors, i.e: O is red, N is blue, C is cyan and S is yellow.
Name. In this coloring method, if you choose a drawing method that shows individual atoms, you can see that they have different colors, i.e: O is red, N is blue, C is cyan and S is yellow.
- 11
- Choose Coloring Method
 ResType (Fig. 7(c)).
This allows you to distinguish non-polar residues (white),
basic residues (blue), acidic residues (red) and polar residues (green).
ResType (Fig. 7(c)).
This allows you to distinguish non-polar residues (white),
basic residues (blue), acidic residues (red) and polar residues (green).
- 12
- Select Coloring Method
 Secondary Structure (Fig. 7(c)) and
confirm that the NewCartoon representation displays colors consistent with secondary
structure.
Secondary Structure (Fig. 7(c)) and
confirm that the NewCartoon representation displays colors consistent with secondary
structure.
You can also only display parts of the molecule that you are interested in by specifying you selection in the Graphical Representations
window (Fig. 7(f)).
Figure 9:
Graphical Representations window and the Selections tab.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_GrapRepr2}
}
\end{center}
\end{figure}](img21.gif) |
- 13
- In the Graphical Representations
window, there is a Selected Atoms text entry (Fig. 7(f)). Delete the word all,
type helix and press the Apply button or hit the Enter/Return key on your keyboard (remember to do this whenever you change a selection).
VMD will show just the helices present in our molecule.
- 14
- In the Graphical Representations window choose
the Selections tab (Fig. 9(a)).
In section Singlewords (Fig. 9(b)), you
will find a list of possible selections you can type. For instance,
try to display
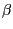 -sheets instead of helices by typing the appropriate
word in the Selected Atoms text entry.
-sheets instead of helices by typing the appropriate
word in the Selected Atoms text entry.
Combinations of boolean operators can also be used when writing a selection.
- 15
- In order to see the molecule without helices and
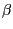 -sheets, type the
following in Selected Atoms: (not helix) and (not betasheet). Remember to press the Apply button or hit the Enter/Return key on your keyboard.
-sheets, type the
following in Selected Atoms: (not helix) and (not betasheet). Remember to press the Apply button or hit the Enter/Return key on your keyboard.
- 16
- In the section Keyword (Fig. 9(c))
of the Selections tab, you can see properties that can be used to select parts of a
protein with their possible values. Look at possible values of
the keyword resname (Fig. 9(d)). Display all the
lysines and glycines present in the
protein by typing (resname LYS) or (resname GLY) in the Selected Atoms. Lysines
play a fundamental role in the configuration of polyubiquitin chains.
- 17
- Now, change the current representation's Drawing Method to CPK
and the Coloring Method to ResName in the Draw Style tab. In the
screen you will be able to see the different Lysines and Glycines.
- 18
- In the Selected
Atoms text entry type water. Choose Coloring Method
 Name. You should see the 58 water molecules
(in fact only the oxygens) present in our system.
Name. You should see the 58 water molecules
(in fact only the oxygens) present in our system.
- 19
- In order to see which water molecules are closer to the protein
you can use the command within. Type water and within 3 of protein for Selected Atoms.
This selects all the water molecules that are within a distance of 3
angstroms of the protein.
- 20
- Finally, try typing the following selections in Selected Atoms:
Table 1:
Example atom selections.
| Selection |
Action |
| protein |
Shows the Protein |
| resid 1 |
The first residue |
| (resid 1 76) and (not water) |
The first and last residues |
| (resid 23 to 34) and (protein) |
The 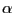 -helix
-helix |
|
The button Create Rep (Fig. 10(a)) in the Graphical Representations window allows you to create multiple representations. Therefore, you can have a mixture of different selections with different styles and colors, all displayed at the same time.
- 21
- For the current representation, in Selected Atoms type protein, set the Drawing Method to NewCartoon
and the Coloring Method to Secondary Structure.
- 22
- Press the Create Rep button (Fig. 10(a)). You should see that a new representation is created.
Modify the new representation to get VDW as the Drawing Method, ResType as the Coloring Method, and resname LYS as the current selection.
- 23
- Repeating the previous procedure, create the following two new
representations:
Table 2:
Example representations.
| Selection |
Coloring Method |
Drawing Method |
| water |
Name |
CPK |
| resid 1 76 and name CA |
ColorID
 1
1 |
VDW |
|
Figure 10:
Multiple Representations of Ubiquitin.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_MultRepr}
}
\end{center}
\end{figure}](img23.gif) |
- 24
- Create the last representation by pressing again the Create Rep button. Select Drawing Method
 QuickSurf for drawing method, Coloring Method
QuickSurf for drawing method, Coloring Method
 Molecule for coloring method, and type protein in the Selected Atoms entry. Set the QuickSurf radius scale parameter to 0.7. For this last representation choose Transparent in the Material pull-down menu (Fig. 10(c)). This representation shows protein's volumetric surface in transparent.
Molecule for coloring method, and type protein in the Selected Atoms entry. Set the QuickSurf radius scale parameter to 0.7. For this last representation choose Transparent in the Material pull-down menu (Fig. 10(c)). This representation shows protein's volumetric surface in transparent.
- 25
- Note that you can select and modify different
representations you have created by clicking on a representation to highlight it in yellow.
Also, you can switch each representation on/off by double-clicking on it. You can also delete a representation by highlighting it and clicking on the Delete Rep button (Fig. 10(b)). At the end
of this section, your Graphical Representations window
should look similar to Fig. 10.
When dealing with a protein for the first time, it is very
useful to find and display different amino acids quickly. The
sequence viewer extension allows you to view the protein sequence, as well as picking and displaying one or more residues of your choice easily.
Figure 11:
VMD Sequence window.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_Seq}
}
\end{center}
\end{figure}](img24.gif) |
- 1
- In the VMD Main window, choose the Extensions
 Analysis
Analysis
 Sequence Viewer menu item.
A window (Fig. 11(a)) with a list of the amino acids (Fig. 11(e)) and their properties (Fig. 11(b)&(c)) will appear in your screen.
Sequence Viewer menu item.
A window (Fig. 11(a)) with a list of the amino acids (Fig. 11(e)) and their properties (Fig. 11(b)&(c)) will appear in your screen.
- 2
- With the mouse, try clicking on different residues in the list (Fig. 11(e)) and
see how they are highlighted. In addition, the highlighted residue will
appear in your OpenGL Display window in yellow and bond
drawing method, so you can visualize its location within the protein easily.
- 3
- Using the Zoom controls (Fig. 11(f)) you can display the entire
list of residues in the window. This is especially useful for
larger proteins
- 4
- To pick multiple residues, hold the shift key and click on the mouse button. Try highlighting residues 11, 48, 63 and 29
(Fig. 11(e)).
- 5
- Look at the Graphical Representations window, you should find a new representation
with the residues you have selected using Sequence Viewer Extension. You can modify, hide or delete this representation similar to what you have done before.
Information about residues is color-coded (Fig. 11(d)) in columns and obtained from STRIDE. The
B-value column (Fig. 11(b)) shows the B-value field (temperature
factor). The struct column shows secondary structure (Fig. 11(d)), where each letter
means:
Table 3:
Secondary Structure codes used by STRIDE.
| T |
Turn |
| E |
Extended conformation (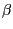 -sheets)
-sheets) |
| B |
Isolated bridge |
| H |
Alpha helix |
| G |
3-10 helix |
| I |
Pi helix |
| C |
Coil |
|
Saving Your Work
The viewpoints and representations that you have created using VMD can be saved as a VMD state. This
VMD state contains all the information needed to reproduce the same VMD session without losing what you have done.
- 1
- Go to the OpenGL Display window, use your mouse to find a nice view of the protein. We will save this viewpoint using VMD ViewMaster.
- 2
- In the VMD Main window, select Extension
 Visualization
Visualization
 ViewMaster. This will open the VMD ViewMaster window.
ViewMaster. This will open the VMD ViewMaster window.
- 3
- In the VMD ViewMaster window, click on the Create New button. Now you have saved your OpenGL Display view point.
- 4
- Go back to your OpenGL Display window, use your mouse to find another nice view. If you want you can also add/delete/modify a representation in the Graphical Representations window. When you have found a good view, you can again save it by returning to the VMD ViewMaster window and clicking on the Creat New button.
- 5
- Create as many views as you like by repeating the previous step. You can see that in the VMD ViewMaster window, all of your viewpoints are displayed as thumbnails. You can go to a previously-saved viewpoint by clicking on its thumbnail.
- 6
- Let's now save the entire VMD session. In the VMD Main window, choose the File
 Save Visualization State menu item.
Write an appropriate name (e.g., myfirststate.vmd) and save it. The VMD state file myfirststate.vmd contains all the information you need to restore your VMD session, including the viewpoints and the representations.
Save Visualization State menu item.
Write an appropriate name (e.g., myfirststate.vmd) and save it. The VMD state file myfirststate.vmd contains all the information you need to restore your VMD session, including the viewpoints and the representations.
To load a saved VMD state, start a new VMD session and in the VMD Main window choose File
 Load State.
Load State.
- 7
- Quit VMD.
One of VMD's many strengths is its ability to render high-resolution, publication-quality molecule images. In this section we will introduce some basic concepts of figure rendering in VMD.
Before you render a figure, you want to make sure you set up the OpenGL Display background the way you want. Nearly all aspects of the OpenGL Display are user-adjustable, including background color.
- 1
- Start a new VMD session. Load the 1ubq.pdb file in the vmd-tutorial-files directory by following the steps in Section 1.1.
- 2
- In the VMD Main window, choose Graphics
 Colors.... The Color Controls window should show up. Look through the Categories
list. All display colors, for example, the colors of different atoms when
colored by name, are set here.
Colors.... The Color Controls window should show up. Look through the Categories
list. All display colors, for example, the colors of different atoms when
colored by name, are set here.
- 3
- Now we will change the background color. In Categories, select Display. In Names, select
Background. Finally, choose 8 white in Colors. Your OpenGL Display now should have a white background.
- 4
- When making a figure, we often don't want to include the axes. To turn off the axes, select Display
 Axes
Axes
 Off in the VMD Main window.
Off in the VMD Main window.
Increasing resolution
Most VMD graphical representations are drawn with an
adjustable resolution, allowing users to balance geometric
detail with interactive drawing speed.
- 5
- Open the Graphical Representation window via Graphics
 Representations... in the VMD Main menu. Modify the default representation to show just the protein, and display it using the VDW drawing method.
Representations... in the VMD Main menu. Modify the default representation to show just the protein, and display it using the VDW drawing method.
- 6
- Zoom in on one or two of the atoms, either by using the scroll
wheel on your mouse, or by using Mouse
 Scale
Mode (shortcut s).
Scale
Mode (shortcut s).
- 7
- Notice that with the default resolution setting, the
``spherical'' atoms aren't looking very spherical. In the
Graphical Representations window, click on the representation you set
up before for the protein to highlight it in yellow. Try
adjusting the Sphere Resolution setting to something higher,
and see what a difference it can make. (See Fig. 12.)
Many of the drawing methods have a resolution setting. Try a
few different drawing methods and see how you can easily increase their resolutions. When producing images, you can raise the
resolution until it stops making a visible difference.
Figure 12:
The effect of the resolution setting.
[Low resolution: Sphere Resolution set to 8] ![\includegraphics[scale=0.20]{FIGS/u2-lowres}](img27.gif)
[High resolution: Sphere Resolution set to 28] ![\includegraphics[scale=0.20]{FIGS/u2-highres}](img28.gif)
|
- 8
- You may have noticed the Material menu in the
Graphical Representations window (which by default is drawn with Opaque material). Choose the protein representation
you made before, and experiment with the different materials in the Material menu.
- 9
- Besides the pre-defined materials in the Material menu, VMD also allows users to create their own materials. To make a new material, in the VMD Main window choose Graphics
 Materials.... In the Materials window that
appears, you'll see a list of the materials you just tried out, and
their adjustable settings. Click the Create New button. A new material, e.g. Material23, will be created. Give it the following settings:
Materials.... In the Materials window that
appears, you'll see a list of the materials you just tried out, and
their adjustable settings. Click the Create New button. A new material, e.g. Material23, will be created. Give it the following settings:
Table 4:
Example user-defined material.
| Setting |
Value |
| Ambient |
0.30 |
| Diffuse |
0.30 |
| Specular |
0.90 |
| Shininess |
0.50 |
| Opacity |
0.95 |
|
- 10
- Go back to the Graphical Representations window. In the Material menu, you can see that Material23 is now on the list. Try using Material23 for a representation and see what it looks like.
Figure 13:
Examples of different material settings.
[The default transparent material.] ![\includegraphics[scale=0.5]{FIGS/u2-trans-2}](img30.gif)
[A user-defined material.] ![\includegraphics[scale=0.5]{FIGS/u2-usertrans-2}](img31.gif)
|
- 11
- If your computer supports GLSL Render Mode, you can try to reproduce Fig. 13(b). First turn on the GLSL rendering mode by selecting Display
 Rendermode
Rendermode
 GLSL in the VMD Main window.
GLSL in the VMD Main window.
- 12
- Modify Material23 to be more transparent by entering the following values in the Materials window:
Table 5:
Example of a more transparent material.
| Setting |
Value |
| Ambient |
0.30 |
| Diffuse |
0.50 |
| Specular |
0.87 |
| Shininess |
0.85 |
| Opacity |
0.11 |
|
- 13
- Hide all of your current representations and create the following two representations:
Table 6:
Example of representations drawn with different materials.
| Selection |
Coloring Method |
Drawing Method |
Material |
| protein |
Structure |
NewCartoon |
Opaque |
| protein |
ColorID
 8 white
8 white |
QuickSurf |
Material23 |
|
Since the systems we are dealing with are three-dimensional,
VMD has multiple ways of representing the third dimension. In this
section, we will explore how to use VMD to enhance or hide
depth perception.
- 14
- The first thing to consider is the projection mode. In the
VMD Main window, click the Display menu. Here we can
choose either Perspective or Orthographic in the drop-down menu. Try switching between Perspective and Orthographic projection modes and see
the difference (Fig. 14).
In perspective mode, things nearer the camera appear larger. Although
perspective projection provides strong size-based visual depth cues, the
displayed image will not preserve scale relationships or parallelism of lines,
and objects very close to the camera may appear distorted.
Orthographic projection preserves scale and parallelism relationships
between objects in the displayed image, but greatly reduces depth perception.
Figure 14:
Comparison of perspective and orthographic projection modes.
[Perspective] ![\includegraphics[scale=0.24]{FIGS/u2-persp}](img32.gif)
[Orthographic] ![\includegraphics[scale=0.24]{FIGS/u2-ortho}](img33.gif)
|
Another way VMD can represent depth is through the so-called
``depth cueing''. Depth cueing is used to enhance three-dimensional
perception of molecular structures, particularly with orthographic
projections.
- 15
- Choose Display
 Depth Cueing in
the VMD Main window. When depth cueing is enabled, objects
further from the camera are blended into the background. Depth cueing
settings are found in Display
Depth Cueing in
the VMD Main window. When depth cueing is enabled, objects
further from the camera are blended into the background. Depth cueing
settings are found in Display
 Display
Settings.... Here you can choose the functional dependence of the
shading on distance, as well as some parameters for this function. To see the effect better, you might want to hide the representation with the QuickSurf drawing method.
Display
Settings.... Here you can choose the functional dependence of the
shading on distance, as well as some parameters for this function. To see the effect better, you might want to hide the representation with the QuickSurf drawing method.
- 16
- Finally, VMD can also produce stereo images. In the VMD
Main window, look at the Display
 Stereo menu,
showing many different choices. Choose SideBySide (remember to return
to Perspective mode for a better result). You should get something like Fig. 15
Stereo menu,
showing many different choices. Choose SideBySide (remember to return
to Perspective mode for a better result). You should get something like Fig. 15
- 17
- Turn off stereo image by selecting Display
 Stereo
Stereo
 Off in the VMD Main window. Also turn off depth cueing by unselecting the Display
Off in the VMD Main window. Also turn off depth cueing by unselecting the Display
 Depth Cueing checkbox in
the VMD Main window.
Depth Cueing checkbox in
the VMD Main window.
Figure 15:
Stereo image of the ubiquitin protein. Shown here with Cue Mode = Linear,
Cue Start = 1.5, and Cue End = 2.75.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=0.5\textwidth]{FIGS/u2-stereo}
}
\end{center}
\end{figure}](img35.gif) |
Rendering
By now we've seen some techniques for producing nice views and representations of the molecule loaded in VMD. Now, we'll explore the use of the VMD built-in snapshot feature and external rendering programs to produce high quality images of your molecule. The ``snapshot''
renderer saves the on-screen image in your OpenGL window and is
often adequate for use in drafts of presentations, movies, and small figures.
When one desires higher quality images for publication,
renderers such as Tachyon (and its hardware-accelerated variants) and
POV-Ray are much better choices due to their improved rendering quality
and support for advanced lighting and shading. One of the key benefits
of Tachyon and POV-Ray vs. the ``snapshot'' renderer is that they can
directly render curved geometric primitives such as spheres and cylinders,
eliminating the need to be concerned with the ``resolution'' parameters
for most graphical representations as described in Sec. 1.6.2.
Sophisticated renderers such as Tachyon and POV-Ray perform challenging
rendering calculations in exchange for the high quality images they produce.
When rendering very high resolution images in combination with large
numbers of transparent surfaces, shadows, ambient occlusion lighting, and
other advanced features, the rendering time required by Tachyon or POV-Ray can
start to become a noteworthy consideration, particularly in the context
of movie rendering. VMD includes an I/O optimized Tachyon rendering
path called TachyonInternal that completely avoids writing VMD
scene files and constituent geometry to disk, instead rendering directly
from the in-memory VMD scene and writing out only the final rendered image.
The use of TachyonInternal provides a large performance benefit when
rendering large geometrically complex VMD scenes and is the basis for
even higher performance hardware-optimized Tachyon variants.
VMD optionally incorporates two hardware-optimized ``light weight'' versions of
the Tachyon rendering engine that use GPU-acceleration and/or CPU vectorization
to achieve higher performance than the fully-general cross-platform
version of Tachyon. These hardware-optimized variants of Tachyon are
referred to as
TachyonLOptiX and TachyonLOptiXInternal (NVIDIA GPU-acceleration), and
TachyonLOSPRay and TachyonLOSPRayInternal (Intel CPU vectorization),
each of which offer substantial performance gains
(frequently 2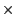 up to as much as 10
up to as much as 10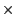 faster)
over the full-featured cross-platform Tachyon renderer.
faster)
over the full-featured cross-platform Tachyon renderer.
- 18
- Hide or delete all your previous representations, and create the new representations shown in Table 7.
Table 7:
Example representations.
| Selection |
Coloring Method |
Drawing Style |
Material |
| protein and not resid 72 to 76 |
Structure |
NewCartoon |
Opaque |
| protein and helix and name CA |
ColorID
 8
8 |
QuickSurf |
Material 23 |
| |
|
|
(or Material |
| |
|
|
with another number) |
| resname GLY and not resid 72 to 76 |
ColorID
 7
7 |
VDW |
Opaque |
| resname LYS |
ColorID
 18
18 |
Licorice |
Opaque |
|
- 19
- Rendering is very simple in VMD. Once you have the scene set the
way you like it in the OpenGL window, simply choose File
 Render... in
the VMD Main window. The File Render Controls
window will appear on your screen.
Render... in
the VMD Main window. The File Render Controls
window will appear on your screen.
- 20
- The File Render Controls allows you to choose which renderer you want to use and the
file name for your image. For our first try, let's select snapshot for the rendering method, type in a filename of your choice, and click Start Rendering.
- 21
- If you are using a Mac or a Linux machine, an image-processing application might open automatically that shows you the molecule you have just rendered using snapshot. If this is not the case, use any image-processing application to take a look at the image file. Close the application when you are done to continue using VMD.
- 22
- Try to render again using different rendering method, particularly
TachyonInternal and POV3. If you're using a GPU-accelerated hardware
platform, try using one of the TachyonLOptiX renderers, and if running
on appropriate Intel CPU hardware, try TachyonLOSPRay.
Compare the quality of the images created by different renderers.
Figure 16:
Example of a POV-Ray 3.6 (POV3) rendering.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=0.35\textwidth]{FIGS/u2-render-hires}
}
\end{center}
\end{figure}](img38.gif) |
- 23
- You have learned the basics of VMD. Quit VMD.


Next: Trajectories and Movie Making
Up: VMD Tutorial
Previous: Introduction
vmd@ks.uiuc.edu
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_MoleFileBrow}
}
\end{center}
\end{figure}](img9.gif)
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{FIGS/u2_rotations}
}
\end{center}
\end{figure}](img12.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
... keys instead of the {\sf Mouse} menu to change the mouse mode.}
\end{minipage}}](img15.gif)
![]() Center menu item. It allows you to specify the point around which rotations are done.
Center menu item. It allows you to specify the point around which rotations are done.
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
...linder, but the sphere radius cannot be modified independently.}
\end{minipage}}](img19.gif)
![]() -sheets as solid
arrows and all other structures as a tube. This is probably the most popular
drawing method to view the overall architecture of a protein.
-sheets as solid
arrows and all other structures as a tube. This is probably the most popular
drawing method to view the overall architecture of a protein.
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
...e the secondary structure according to an heuristic algorithm.
}
\end{minipage}}](img20.gif)
![]() Load State.
Load State.
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
...te the camera into individual atoms
without clipping them off.
}
\end{minipage}}](img26.gif)
![\includegraphics[scale=0.20]{FIGS/u2-lowres}](img27.gif) [High resolution: Sphere Resolution set to 28]
[High resolution: Sphere Resolution set to 28]![\includegraphics[scale=0.20]{FIGS/u2-highres}](img28.gif)
![% latex2html id marker 3317
\framebox[\textwidth]{
\begin{minipage}{.2\textwidt...
...-blended transparency; see
Fig.~\ref{fig:trans} for an example.}
\end{minipage}}](img29.gif)
![\includegraphics[scale=0.5]{FIGS/u2-trans-2}](img30.gif) [A user-defined material.]
[A user-defined material.]![\includegraphics[scale=0.5]{FIGS/u2-usertrans-2}](img31.gif)
![\includegraphics[scale=0.24]{FIGS/u2-persp}](img32.gif) [Orthographic]
[Orthographic]![\includegraphics[scale=0.24]{FIGS/u2-ortho}](img33.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
...ive mode is often used for producing figures and
stereo images.}
\end{minipage}}](img34.gif)
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=0.5\textwidth]{FIGS/u2-stereo}
}
\end{center}
\end{figure}](img35.gif)
![]() up to as much as 10
up to as much as 10![]() faster)
over the full-featured cross-platform Tachyon renderer.
faster)
over the full-featured cross-platform Tachyon renderer.
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
....01 to get a better idea of what will
appear in your rendering.}
\end{minipage}}](img37.gif)