


Next: Investigating Structural Alignment
Up: Aquaporin Tutorial
Previous: Comparing Protein Sequence and
Contents
Subsections
Residue Selection
In this section, you will use the Select Residues tool for
finding structural features of aquaporins that have been conserved throughout
species and are crucial for their function.
The Select Residues features analysis of
structural similarity and sequence conservation. It makes use of different measures to highlight residues
in the Sequence Display and Structure Display simultaneously. It allows you to examine the conservation and similarity on
a per residue basis, using two different measures, Qres value measuring structure
similarity and Sequence Identity measuring sequence conservation.
You can use Multiseq to look at the residues that are structurally similar above or below a certain value of Qres.
Figure:
Residue Selection
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.2 in]{pictures/rsmenu}
}
\end{center}\vspace{-0.7cm}\end{figure}](img36.gif) |
- 1
- Choose the Search
 Select Residues tool. A
side menu will appear (Fig. 17). Within the
menu, you can select residues based on their Sequence
Identity or Qres.
Select Residues tool. A
side menu will appear (Fig. 17). Within the
menu, you can select residues based on their Sequence
Identity or Qres.
- 2
- In box following Qres, select
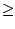 and key in 0.5 for the value.
and key in 0.5 for the value.
- 3
- Click on the Select button.
Note the changes in the display of the Open GL (structure) Display
window (Fig. 18, left) and in the Sequence Display of the Multiseq program window
(Fig. 19).
- 4
- Look in the Sequence Display
(Fig. 19). Since you selected the Qres value to be greater or
equal to 0.5, most of the structural elements of the aligned molecules will be
shown as conserved (similar), and will be displayed in yellow.
Figure:
View of the Residue Selection for Qres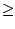 0.5 in the Open GL
Display window. Highlight style set to Licorice (left) and NewCartoon (right)
0.5 in the Open GL
Display window. Highlight style set to Licorice (left) and NewCartoon (right)
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=4 in]{pictures/bulkbyq-05}
}
\end{center}\vspace{-0.7cm}\end{figure}](img37.gif) |
- 5
- Now, look at the OpenGL Display window (Fig. 18,
left). In this case also
most parts of the molecules are highlighted, in particular, the transmembrane segments.
Figure:
View of the Residue Selection for Q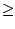 0.5 in
the Sequence Display window.
0.5 in
the Sequence Display window.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.5 in]{pictures/bulkseqbyq-05}
}
\end{center}\vspace{-0.7cm}\end{figure}](img38.gif) |
- 6
- Due to the high level of conservation, the Licorice highlighting
style is not very informative. To improve the view, chose the View
 Highlight Style menu item, and click on the New Cartoon
style (Fig. 18, right). You can recognize immediately that
high Qres values indicating conservation of neighborhood relationships for amino
acids are seen in the transmembrane region throughout.
Highlight Style menu item, and click on the New Cartoon
style (Fig. 18, right). You can recognize immediately that
high Qres values indicating conservation of neighborhood relationships for amino
acids are seen in the transmembrane region throughout.
Now you will investigate what happens for higher values of Qres.
- 7
- In the Select Residues window, select
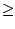 after the Qres and key in 0.75.
Click on the Select button.
after the Qres and key in 0.75.
Click on the Select button.
- 8
- Look at the OpenGL Display window
(Fig. 20). Notice that most of the structurally
similar regions
are still in the transmembrane region, but the highlighted regions are
more confined than before.
Figure:
View of the Residue Selection for Qres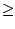 0.75 in
the OpenGL Display window
0.75 in
the OpenGL Display window
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.0 in]{pictures/bulkbyq-07}
}
\end{center}\vspace{-0.7cm}\end{figure}](img39.gif) |
- 9
- Look in the Sequence Display window
(Fig. 21). You can still distinguish blocks of
structurally similar residues, which correspond to the helices. They are
not as well defined as before, but the structural similarity is still
noticeable when probing for high values of Qres.
Figure:
View of the Residue Selection for Qres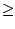 0.75 in
the Sequence Display window
0.75 in
the Sequence Display window
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.5 in]{pictures/bulkseqbyq-07}
}
\end{center}\vspace{-0.7cm}\end{figure}](img40.gif) |
- 10
- Repeat steps 7 to 9 and set the Qres to
 0.9. You will notice that almost all the structure
conservation is lost.
0.9. You will notice that almost all the structure
conservation is lost.
What does this mean? The aligned aquaporins have a moderate to high
structure conservation. Structurally, the most similar areas are the
transmembrane helices.
You can also use Select Residues to look at the residues that are conserved on the
sequence level.
- 1
- Go back to the main Multiseq program window and select Search
 Select Residues.
Select Residues.
Figure:
View of the Residue Selection by Sequence
Identity in the OpenGL Display window.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.7 in]{pictures/bulkbyseqid}
}
\end{center}\vspace{-0.7cm}\end{figure}](img41.gif) |
- 2
- In the Select Residues window, following Sequence Identity select
 .
.
- 3
- You can then key in a value between 0 and 100. Try 70, which will select residues that were assigned a
sequence identity measure of 70% or more.
- 4
- Click on the Select button.
Figure:
View of the Residue Selection by Sequence
Identity in the Sequence Display window
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.5 in]{pictures/bulkseqbyseqid}
}
\end{center}\vspace{-0.7cm}\end{figure}](img42.gif) |
- 5
- Look at the Sequence Display in the Multiseq program window. You will notice very few
residues are selected. If you look carefuly, you will locate some of
the residues that correspond to key structural features of aquaporins, for
which you made representations in section 1 of this tutorial. Search
for the NPA motifs, highlighted in yellow (Fig. 23).
- 6
- In the OpenGL window, look at the areas highlighted in
yellow. Notice that many residues in the reentrant loops (Fig. 3) conserved their
sequence over the course of evolution. This is a clear indication that the
reentrant loops are functionally significant.
The conserved amino acids in the sequence are mostly located in the pore, and
in the reentrant loops. You will now compare these residues to the
representations you created in section 1 of this tutorial, which correspond to
residues that are relevant to the function of aquaporins.
- 7
- In the Multiseq program window, select View
 Highlight Style
Highlight Style  Licorice.
Licorice.
- 8
- In the VMD Main window, select Graphics
 Representations....
Representations....
- 9
- In the Graphical Representations window, in the Selected
Molecules pull-down menu, select the molecule 1j4n.
- 10
- In the Graphical Representations window, double click on the
representations you created for the NPA motif, the glutamates and the
arginine. What is the sequence identity for these amino acids? Why do you think the NPA
motif, the lysine and the glutamates are conserved throughout species?
When you are done examining the NPA motif, close the Graphical Representations window
to proceed to the next section.



Next: Investigating Structural Alignment
Up: Aquaporin Tutorial
Previous: Comparing Protein Sequence and
Contents
school@ks.uiuc.edu
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=2.2 in]{pictures/rsmenu}
}
\end{center}\vspace{-0.7cm}\end{figure}](img36.gif)
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=4 in]{pictures/bulkbyq-05}
}
\end{center}\vspace{-0.7cm}\end{figure}](img37.gif)