

Next: Ubiquitin in a Water
Up: Basics of NAMD
Previous: Generating a Protein Structure
Subsections
Now, the protein needs to be solvated, i.e., put inside water, to more closely resemble the cellular environment. You will do so in two ways, placing ubiquitin in:
- a water sphere in surrounding vacuum, in preparation for minimization and equilibration without periodic boundary conditions.
- a water box, in preparation for minimization and equilibration with periodic boundary conditions.
You will utilize a prepared tcl script to create the water sphere. It is called wat_sphere.tcl and is located in your 1-1-build directory. The syntax of the script itself is given in Appendix G.
- 1
- In a Terminal window in your 1-1-build directory, type
vmd -dispdev text -e wat_sphere.tcl
|
|
This will call the script, which will place ubiquitin in the smallest possible water sphere which completely immerses the protein.
- 2
- The output of the wat_sphere.tcl script will be the center and radius of the water sphere. Record these numbers. You will need them later. The script will have created the pdb and psf files ubq_ws.pdb and ubq_ws.psf, respectively, of ubiquitin in a water sphere. They will be in your 1-1-build directory. Check this by typing ls .
- 3
- Type exit in the Terminal to exit the text version of VMD.
- 4
- Launch the graphical version of VMD by typing vmd in the Terminal window.
We will check the appearance of the immersed molecule by loading the molecule with both structural and coordinate information.
- 5
- Load the structural information first. Click File
 New Molecule... in the VMD Main window. In the Molecule File Browser use the Browse... button to find the file ubq_ws.psf. Load it by pressing the Load button.
New Molecule... in the VMD Main window. In the Molecule File Browser use the Browse... button to find the file ubq_ws.psf. Load it by pressing the Load button.
- 6
- Load the coordinates into the PSF file. Make sure the Load files for: field in the Molecule File Browser says ubq_ws.psf. Then, click Browse
 , find the file ubq_ws.pdb, and load it by pressing the Load button.
, find the file ubq_ws.pdb, and load it by pressing the Load button.
You should be able to see your ubiquitin molecule in the center of a beautiful sphere of water. Changing the representations in VMD will enable you to see it better.
- 7
- Delete your ubiquitin molecule in a water sphere using the VMD Main window by clicking on ubq_ws.psf in the window, and selecting Molecule
 Delete Molecule.
Delete Molecule.
- 1
- In the VMD Main window, click Extensions
 Tk Console. In the VMD TkCon window of your VMD session type:
Tk Console. In the VMD TkCon window of your VMD session type:
| package require solvate |
|
solvate ubq.psf ubq.pdb -t 5 -o ubq_wb
|
|
The package require solvate command loads the solvate package, so that VMD will be able to call it.
The solvate package will put your protein (described in ubq.psf and ubq.pdb) in a box of water. The -t option creates the water box dimensions such that there is a layer of water 5Å in each direction from the atom with the largest coordinate in that direction. The -o option creates the output files ubq_wb.pdb and ubq_wb.psf for ubiquitin with the water box. You will also obtain the files combine.pdb and combine.psf which may be eliminated. They are intermediate files used by the solvate program to transport information into VMD.
- 2
- In the VMD Main window, click File
 New Molecule. In the Molecule File Browser use the Browse... button to find the file ubq_wb.psf. Load it by pressing the Load button. This will load the structural information into VMD.
New Molecule. In the Molecule File Browser use the Browse... button to find the file ubq_wb.psf. Load it by pressing the Load button. This will load the structural information into VMD.
- 3
- In the same Molecule File Browser, browse for ubq_wb.pdb and load it. In the display, check if the water box surrounds ubiquitin, and demonstrate to yourself that the hydrogen atoms now exist.
- 4
- In the VMD TkCon window type:
| set everyone [atomselect top all] |
|
measure minmax $everyone
|
|
This analyzes all atoms in the system and gives you the minimum and maximum values of 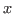 ,
, 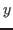 and
and 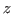 coordinates of the entire protein-water system.
coordinates of the entire protein-water system.
- 5
- These values are defined relative to the origin of the coordinate system,
set by the initial pdb file, 1UBQ.pdb. The center of the water box may
be determined by calculating the midpoint of each of the three sides in the
coordinate system. For example, if minimum and maximum values of
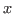 are
returned as 10.44 and 51.12, respectively,
are
returned as 10.44 and 51.12, respectively, 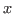 -coordinate of the center of the
box would be
-coordinate of the center of the
box would be
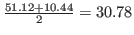 . Alternatively, one can use tcl
scripting in VMD to find the center of the box by typing
. Alternatively, one can use tcl
scripting in VMD to find the center of the box by typing
in the VMD TkCon window. Determine the coordinates of the center of your water box and record these numbers.
- 6
- Close VMD by choosing File
 Quit in the VMD Main window.
Quit in the VMD Main window.
Figure 3:
Ubiquitin in a water box and in a water sphere. Hydrogen atoms are colored black for contrast.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{pictures/tut_unit01_wb_ws}
}
\end{center}
\end{figure}](img34.png) |
Now that you have the pdb and psf files for solvated ubiquitin, along with the parameter file for proteins in general, you only need a NAMD configuration file to be able to minimize and equilibrate your ubiquitin-water system. In the next two sections, you will edit a NAMD configuration file provided to you in order to gain a beginner's understanding of its function.
- 7
- Before beginning the next section, you must copy the necessary pdb and psf files into your common folder. We will store the files there so we have a single directory from which to access them, and so that we do not need to keep multiple copies of them. In a Terminal window, type cp ubq.pdb ubq.psf ubq_w* ../common/ and hit Enter. Then, type ls ../common/ to see which files you have copied.


Next: Ubiquitin in a Water
Up: Basics of NAMD
Previous: Generating a Protein Structure
namd@ks.uiuc.edu

![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...eded to achieve equilibration and saves computational effort.}
\end{minipage} }](img25.png)



![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{pictures/tut_unit01_wb_ws}
}
\end{center}
\end{figure}](img34.png)
