The geometric path collective variables define the progress along a path, ![]() , and the distance from the path,
, and the distance from the path, ![]() . These CVs are proposed by Leines and Ensing[54] , which differ from that[55] proposed by Branduardi et al., and utilize a set of geometric algorithms. The path is defined as a series of frames in the atomic Cartesian coordinate space or the CV space.
. These CVs are proposed by Leines and Ensing[54] , which differ from that[55] proposed by Branduardi et al., and utilize a set of geometric algorithms. The path is defined as a series of frames in the atomic Cartesian coordinate space or the CV space. ![]() and
and ![]() are computed as
are computed as
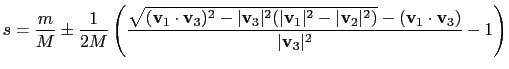 |
(13.13) |
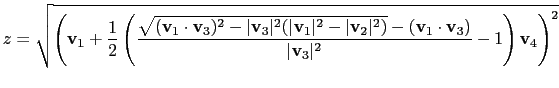 |
(13.14) |
where
![]() is the vector connecting the current position to the closest frame,
is the vector connecting the current position to the closest frame,
![]() is the vector connecting the second closest frame to the current position,
is the vector connecting the second closest frame to the current position,
![]() is the vector connecting the closest frame to the third closest frame, and
is the vector connecting the closest frame to the third closest frame, and
![]() is the vector connecting the second closest frame to the closest frame.
is the vector connecting the second closest frame to the closest frame. ![]() and
and ![]() are the current index of the closest frame and the total number of frames, respectively. If the current position is on the left of the closest reference frame, the
are the current index of the closest frame and the total number of frames, respectively. If the current position is on the left of the closest reference frame, the ![]() in
in ![]() turns to the positive sign. Otherwise it turns to the negative sign.
turns to the positive sign. Otherwise it turns to the negative sign.
The equations above assume: (i) the frames are equidistant and (ii) the second and the third closest frames are neighbouring to the closest frame. When these assumptions are not satisfied, this set of path CV should be used carefully.
In the gspath {...} and the gzpath {...} block all vectors, namely
![]() and
and
![]() are defined in atomic Cartesian coordinate space. More specifically,
are defined in atomic Cartesian coordinate space. More specifically,
![]() , where
, where
![]() is the
is the ![]() -th atom specified in the atoms block.
-th atom specified in the atoms block.
![]() , where
, where
![]() means the
means the ![]() -th atom of the
-th atom of the ![]() -th reference frame.
-th reference frame.
List of keywords (see also ![[*]](crossref.png) for additional options):
for additional options):
List of keywords (see also ![[*]](crossref.png) for additional options):
for additional options):
The usage of gzpath and gspath is illustrated below:
colvar {
# Progress along the path
name gs
# The path is defined by 5 reference frames (from string-00.pdb to string-04.pdb)
# Use atomic coordinate from atoms 1, 2 and 3 to compute the path
gspath {
atoms {atomnumbers { 1 2 3 }}
refPositionsFile1 string-00.pdb
refPositionsFile2 string-01.pdb
refPositionsFile3 string-02.pdb
refPositionsFile4 string-03.pdb
refPositionsFile5 string-04.pdb
}
}
colvar {
# Distance from the path
name gz
# The path is defined by 5 reference frames (from string-00.pdb to string-04.pdb)
# Use atomic coordinate from atoms 1, 2 and 3 to compute the path
gzpath {
atoms {atomnumbers { 1 2 3 }}
refPositionsFile1 string-00.pdb
refPositionsFile2 string-01.pdb
refPositionsFile3 string-02.pdb
refPositionsFile4 string-03.pdb
refPositionsFile5 string-04.pdb
}
}
This is a helper CV which can be defined as a linear combination of other CVs. It maybe useful when you want to define the gspathCV {...} and the gzpathCV {...} as combinations of other CVs.
In the gspathCV {...} and the gzpathCV {...} block all vectors, namely
![]() and
and
![]() are defined in CV space. More specifically,
are defined in CV space. More specifically,
![]() , where
, where ![]() is the
is the ![]() -th CV.
-th CV.
![]() , where
, where
![]() means the
means the ![]() -th CV of the
-th CV of the ![]() -th reference frame. It should be note that these two CVs requires the pathFile option, which specifies a path file. Each line in the path file contains a set of space-seperated CV value of the reference frame. The sequence of reference frames matches the sequence of the lines.
-th reference frame. It should be note that these two CVs requires the pathFile option, which specifies a path file. Each line in the path file contains a set of space-seperated CV value of the reference frame. The sequence of reference frames matches the sequence of the lines.
List of keywords (see also ![[*]](crossref.png) for additional options):
for additional options):
List of keywords (see also ![[*]](crossref.png) for additional options):
for additional options):
The usage of gzpathCV and gspathCV is illustrated below:
colvar {
# Progress along the path
name gs
# Path defined by the CV space of two dihedral angles
gspathCV {
pathFile ./path.txt
dihedral {
name 001
group1 {atomNumbers {5}}
group2 {atomNumbers {7}}
group3 {atomNumbers {9}}
group4 {atomNumbers {15}}
}
dihedral {
name 002
group1 {atomNumbers {7}}
group2 {atomNumbers {9}}
group3 {atomNumbers {15}}
group4 {atomNumbers {17}}
}
}
}
colvar {
# Distance from the path
name gz
gzpathCV {
pathFile ./path.txt
dihedral {
name 001
group1 {atomNumbers {5}}
group2 {atomNumbers {7}}
group3 {atomNumbers {9}}
group4 {atomNumbers {15}}
}
dihedral {
name 002
group1 {atomNumbers {7}}
group2 {atomNumbers {9}}
group3 {atomNumbers {15}}
group4 {atomNumbers {17}}
}
}
}