



Next: Distances
Up: Defining collective variables
Previous: Defining collective variables
Contents
Index
In this context, the function that computes a colvar is called a component.
A component's choice and definition consists of including in the variable's configuration a keyword indicating the type of function (e.g. rmsd), followed by a definition block specifying the atoms involved (see ![[*]](crossref.png) ) and any additional parameters (cutoffs, ``reference'' values, ...).
At least one component must be chosen to define a variable: if none of the keywords listed below is found, an error is raised.
) and any additional parameters (cutoffs, ``reference'' values, ...).
At least one component must be chosen to define a variable: if none of the keywords listed below is found, an error is raised.
The following components implement functions with a scalar value (i.e. a real number):
- distance: distance between two groups;
- distanceZ: projection of a distance vector on an axis;
- distanceXY: projection of a distance vector on a plane;
- distanceInv: mean distance between two groups of atoms (e.g. NOE-based distance);
- angle: angle between three groups;
- dihedral: torsional (dihedral) angle between four groups;
- dipoleAngle: angle between two groups and dipole of a third group;
- dipoleMagnitude magnitude of the dipole of a group of atoms;
- polarTheta: polar angle of a group in spherical coordinates;
- polarPhi: azimuthal angle of a group in spherical coordinates;
- coordNum: coordination number between two groups;
- selfCoordNum: coordination number of atoms within a
group;
- hBond: hydrogen bond between two atoms;
- rmsd: root mean square deviation (RMSD) from a set of
reference coordinates;
- eigenvector: projection of the atomic coordinates on a
vector;
- orientationAngle: angle of the best-fit rotation from
a set of reference coordinates;
- orientationProj: cosine of orientationProj;
- spinAngle: projection orthogonal to an axis of the best-fit rotation
from a set of reference coordinates;
- tilt: projection on an axis of the best-fit rotation
from a set of reference coordinates;
- gyration: radius of gyration of a group of atoms;
- inertia: moment of inertia of a group of atoms;
- inertiaZ: moment of inertia of a group of atoms around a chosen axis;
- alpha:
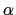 -helix content of a protein segment.
-helix content of a protein segment.
- dihedralPC: projection of protein backbone dihedrals onto a dihedral principal component.
Some components do not return scalar, but vector values:
- distanceVec: distance vector between two groups (length: 3);
- distanceDir: unit vector parallel to distanceVec (length: 3);
- cartesian: vector of atomic Cartesian coordinates (length:
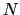 times the number of Cartesian components requested, X, Y or Z);
times the number of Cartesian components requested, X, Y or Z);
- distancePairs: vector of mutual distances (length:
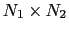 );
);
- orientation: best-fit rotation, expressed as a unit quaternion (length: 4).
The types of components used in a colvar (scalar or not) determine the
properties of that colvar, and particularly which biasing or analysis methods
can be applied.
What if ``X'' is not listed? If a function type is not available on this list, it may be possible to define it as a polynomial superposition of existing ones (see ![[*]](crossref.png) ), or a scripted function (see
), or a scripted function (see ![[*]](crossref.png) ).
).
In the rest of this section, all available component types are listed, along with their physical units and the ranges of values, if limited.
Such limiting values can be used to define lowerBoundary and upperBoundary in the parent colvar.
For each type of component, the available configurations keywords are listed:
when two components share certain keywords, the second component references to
the documentation of the first one that uses that keyword.
The very few keywords that are available for all types of components are listed in a separate section ![[*]](crossref.png) .
.




Next: Distances
Up: Defining collective variables
Previous: Defining collective variables
Contents
Index
vmd@ks.uiuc.edu
![[*]](crossref.png) ) and any additional parameters (cutoffs, ``reference'' values, ...).
At least one component must be chosen to define a variable: if none of the keywords listed below is found, an error is raised.
) and any additional parameters (cutoffs, ``reference'' values, ...).
At least one component must be chosen to define a variable: if none of the keywords listed below is found, an error is raised.
![[*]](crossref.png) ) and any additional parameters (cutoffs, ``reference'' values, ...).
At least one component must be chosen to define a variable: if none of the keywords listed below is found, an error is raised.
) and any additional parameters (cutoffs, ``reference'' values, ...).
At least one component must be chosen to define a variable: if none of the keywords listed below is found, an error is raised.
![[*]](crossref.png) ), or a scripted function (see
), or a scripted function (see ![[*]](crossref.png) ).
).
![[*]](crossref.png) .
.