



Next: Writing a Colvars configuration:
Up: Collective Variables Interface (Colvars)
Previous: Collective Variables Interface (Colvars)
Contents
Index
Subsections
In molecular dynamics simulations, it is often useful to reduce the large number of degrees of freedom of a physical system into few parameters whose statistical distributions can be analyzed individually, or used to define biasing potentials to alter the dynamics of the system in a controlled manner.
These have been called `order parameters', `collective variables', `(surrogate) reaction coordinates', and many other terms.
Here we use primarily the term `collective variable', often shortened to colvar, to indicate any differentiable function of atomic Cartesian coordinates,
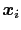 , with
, with 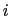 between
between 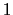 and
and 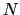 , the total
number of atoms:
, the total
number of atoms:
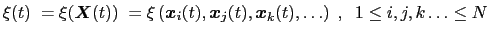 |
(13.1) |
The Colvars module in VMD may be used to calculate these functions over a molecular structure, and to analyze the results of previous simulations.
The module is designed to perform multiple tasks concurrently during or after a simulation, the most common of which are:
- apply restraints or biasing potentials to multiple variables, tailored on the system by choosing from a wide set of basis functions, without limitations on their number or on the number of atoms involved;
- calculate potentials of mean force (PMFs) along any set of variables, using different enhanced sampling methods, such as Adaptive Biasing Force (ABF), metadynamics, steered MD and umbrella sampling; variants of these methods that make use of an ensemble of replicas are supported as well;
- calculate statistical properties of the variables, such as running averages and standard deviations, correlation functions of pairs of variables, and multidimensional histograms: this can be done either at run-time without the need to save very large trajectory files, or after a simulation has been completed using VMD and the cv command.
Note: although restraints and PMF algorithms are primarily used during simulations, they are also available in VMD to test a new input for a simulation, or to evaluate the relative free energy of a new structure based on data from a previous calculation. Options that only have an effect during a simulation are also included for compatibility purposes.
Detailed explanations of the design of the Colvars module are provided in reference [48]. Please cite this reference whenever publishing work that makes use of this module.
Within VMD, the Colvars Module can be accessed in two ways:
- Using the Colvars dashboard, an intuitive, but partial interface to the Colvars module, to easily define and analyze collective variables, but not biases (section
![[*]](crossref.png) ).
).
- Using the full-featured Tcl scripting interface as documented in section
![[*]](crossref.png) ; see in particular the example in section
; see in particular the example in section ![[*]](crossref.png) .
.




Next: Writing a Colvars configuration:
Up: Collective Variables Interface (Colvars)
Previous: Collective Variables Interface (Colvars)
Contents
Index
vmd@ks.uiuc.edu
![]() , with
, with ![]() between
between ![]() and
and ![]() , the total
number of atoms:
, the total
number of atoms:
![[*]](crossref.png) ).
).
![[*]](crossref.png) ; see in particular the example in section
; see in particular the example in section ![[*]](crossref.png) .
.