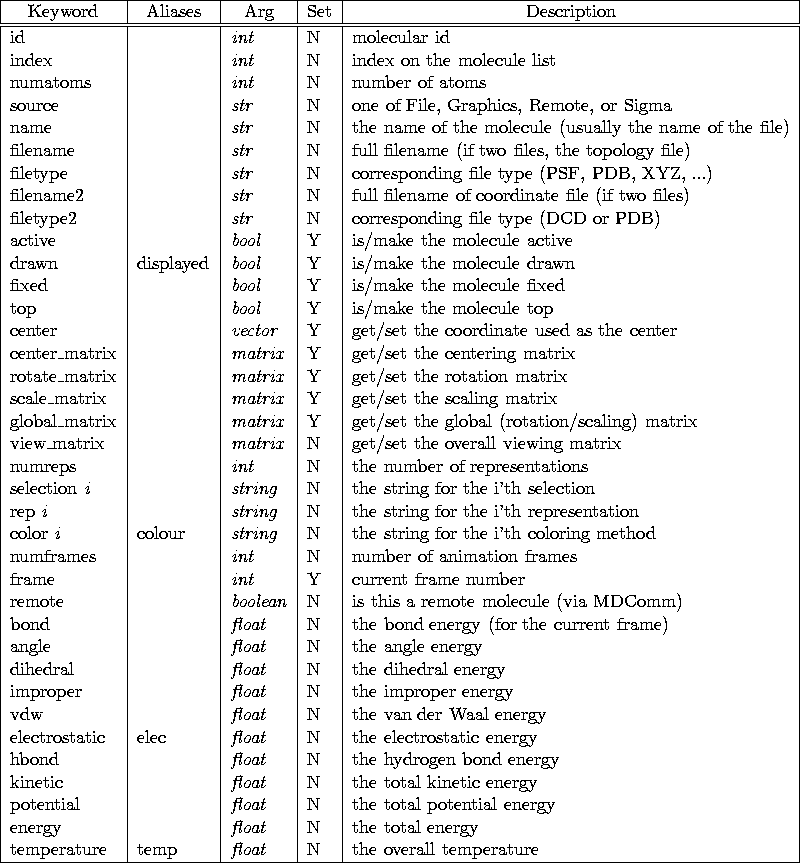
Table 14.1: molinfo keywords
The molinfo command can also be used to access and, in some cases, modify information specific to a given molecule. A query is in the form:
Examples:
vmd > molinfo top get numatoms
Info) 568
vmd > molinfo 0 get {source filename}
Info) File /home/dalke/pdb/pti.pdb
This next example is a bit more complicated. It loops through all the
graphical representation (numreps) and, for each one,
gets the representation used (rep), the selection text
(selection), and the coloring method (color).
vmd > for {set i 0} {$i < [molinfo top get numreps]} {incr i} {
? lassign [molinfo top get "{rep $i} {selection $i} {color $i}"] a b c
? puts "view $i:"
? puts " representation: $a"
? puts " selection: $b"
? puts " coloring method: $c"
? }
view 0:
representation: Tube 0.300000 8.000000
selection: protein backbone
coloring: method Structure
view 1:
representation: Lines 2.000000
selection: same residue as name "S.*"
coloring: method ResName
view 2:
representation: VDW 1.000000 6.000000
selection: name "S.*"
coloring: method Name
A similar behavior can be achieved by using mol list top
,
in which case, however, there is no direct access to the data.
A complete list of keywords is given in Table
14.1.2.
The molinfo command, contrary to its name, can also be used to set some keyword values, such as the current frame number and the display state flags. This duplicates some of the functionality of the mol command, though there are distinct differences in the implementation. Specifically, the mol command uses the internal command queue which, among other things, notifies the appropriate forms that a change occured, redraws the graphics, and logs the commands to the log file, of logging is enabled. In future versions of VMD there will be only one command; for now we suggest only using the molinfo command to get information and to set the frame value and the various viewing matrices.
Examples:
Two functions, one to save the current view position, the other to restore it. The position of the axis is not changed by these operations.
proc save_viewpoint {} {
global viewpoints
if [info exists viewpoints] {unset viewpoints}
# get the current matricies
foreach mol [molinfo list] {
set viewpoints($mol) [molinfo $mol get {
center_matrix rotate_matrix scale_matrix global_matrix}]
}
}
proc restore_viewpoint {} {
global viewpoints
foreach mol [molinfo list] {
puts "Trying $mol"
if [info exists viewpoints($mol)] {
molinfo $mol set {center_matrix rotate_matrix scale_matrix
global_matrix} $viewpoints($mol)
}
}
}
Cycle through the list of displayed molecules, turning each one on one at a time. At the end, return the display flags to their original state.
# save the current display state
foreach mol [molinfo list] {
set disp($mol) [molinfo $mol get drawn]
}
# turn everything off
mol off all
# turn each molecule on then off again
foreach mol [molinfo list] {
if $disp($mol) {
mol on $mol
sleep 1
mol off $mol
}
}
# turn the original ones back on
foreach mol [molinfo list] {
if $disp($mol) {mol on $mol }
}
The last loop, which turns the originally drawn molecules back on, doesn't turn them on at the same time. That's because some commands (those which use the command queue) redraw the graphics when they are used. This can be disabled with the display update (see section § for more information). Using this, the final loop becomes
#turn the original ones back on
display update off
foreach mol [molinfo list] {
if $disp($mol) {mol on $mol }
}
display update on
Alternatively, since the drawn option is settable, you could
do:
foreach mol [molinfo list] {
if $disp($mol) {molinfo $mol set drawn 1}
}
However, that won't set the flag to redraw the scene so you need to force a
redraw with display redraw.