From: Jérôme Hénin (jerome.henin_at_ibpc.fr)
Date: Tue Dec 08 2015 - 09:09:58 CST
Hi "sunyzero",
How many degrees of freedom do you need to sample for your calculation to
work?
Once you can answer that question, you'll have a much better idea how to
proceed.
Jerome
On 8 December 2015 at 08:42, Souvik Sinha <souvik.sinha893_at_gmail.com> wrote:
> I think increasing the sampling time will eventually decrease the barrier
> until convergence of sampling is achieved . And you can also use the
> "historyFreq " option to your restarin1.in file so that you can have the
> track for the pmf during the simulation.
>
> On Tue, Dec 8, 2015 at 11:18 AM, sunyzero <sunyzero_at_163.com> wrote:
>
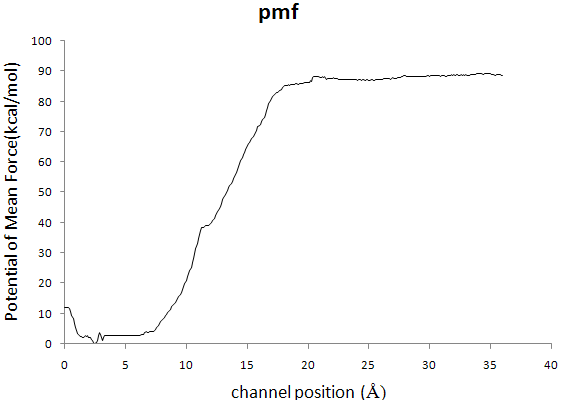
>> Dear Namd users
>> Hello, I need your help about determining the PMF with Adaptive
>> Biasing Forces. According to tutorial about Methods for calculating
>> Poteintials of Mean Force, I use it on HIV-1 protease inhibitors
>> dissociation process,and inhibitors dissociate from binding site moving
>> range from 0 to 36Å, which is divided into six window,each 6-Å wide,but
>> in the ABF output .pmf file, the value of pmf is too large,the maximum
>> value is 88kcal/mol. I try many times, there still exist questions.Here
>> is one of my configures.
>> # Force-Field Parameters
>> exclude scaled1-4
>> 1-4scaling 1.0
>> cutoff 12.
>> switching on
>> switchdist 10.
>> pairlistdist 13.5
>> # Integrator Parameters
>> timestep 1.0 ;# 2fs/step (only if needed to finish quickly)
>> rigidBonds water ;# needed for 2fs steps
>> nonbondedFreq 2
>> fullElectFrequency 4
>> stepspercycle 20
>> # Constant Temperature Control
>> langevin on ;# do langevin dynamicsgmail.google.com
>> langevinDamping 1 ;# damping coefficient (gamma) of 5/ps
>> langevinTemp $temperature
>> langevinHydrogen off ;# don't couple langevin bath to hydrogens
>> wrapAll on
>> # PME (for full-system periodic electrostatics)
>> PME yes
>> PMEGridSpacing 1.0
>> # Constant Pressure Control (variable volume)
>> useGroupPressure no ;# needed for rigidBonds
>> useFlexibleCell no
>> useConstantArea no
>>
>> langevinPiston on
>> langevinPistonTarget 1.01325 ;# in bar -> 1 atm
>> langevinPistonPeriod 100.
>> langevinPistonDecay 50.
>> langevinPistonTemp $temperature
>> # Output
>> outputName win1
>> restartfreq 500 ;# 1000steps = every 1ps
>> dcdfreq 500
>> xstFreq 500
>> outputEnergies 500
>> outputPressure 500
>> colvars on
>> colvarsConfig restrain1.in
>> # Minimization
>> #minimize 500
>> #reinitvels $temperature
>> run 1500000 ;#
>>
>>
>> And here is my restrain1.in
>> colvarsTrajFrequency 500
>> colvarsRestartFrequency 500
>> colvar {
>> name Translocation
>>
>> width 0.1
>>
>> lowerboundary 0.0
>> upperboundary 6.0
>>
>> lowerwallconstant 100.0
>> upperwallconstant 100.0
>>
>> distanceZ {
>> main {
>> atomnumbers { the atoms of ligand }
>> }
>> ref {
>> atomnumbers { the atoms of protein and name CA
>> }
>> }
>> axis ( 0.0, 0.0, 1.0 )
>> }
>> }
>>
>> abf {
>> colvars Translocation
>> fullSamples 1000
>>
>>
>>
>>
>>
>
>
>
> --
> Souvik Sinha
> Research Fellow
> Bioinformatics Centre (SGD LAB)
> Bose Institute
>
> Contact: 033 25693275
>

This archive was generated by hypermail 2.1.6 : Tue Dec 27 2016 - 23:21:37 CST