



Next: Adaptive Tempering
Up: Accelerated Sampling Methods
Previous: Gaussian Accelerated Molecular Dynamics
Contents
Index
Subsections
Solute Scaling and REST2
Solute scaling improves sampling efficiency
by scaling the intramolecular potential energy of a protein
to lower barriers separating different confirmations [79].
The potential is scaled based on a parameter 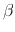 ,
,
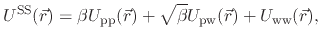 |
(49) |
with
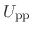 denoting protein-protein interactions,
denoting protein-protein interactions,
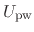 denoting protein-water interactions,
and
denoting protein-water interactions,
and
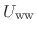 denoting water-water interactions,
effectively ``heating'' the protein's interatomic interactions
whenever
denoting water-water interactions,
effectively ``heating'' the protein's interatomic interactions
whenever 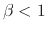 .
The NAMD implementation is made efficient by rescaling
the force field parameters for the affected atoms [31].
In particular, this parameter scaling approach makes the calculation
compatible with existing CUDA force kernels.
.
The NAMD implementation is made efficient by rescaling
the force field parameters for the affected atoms [31].
In particular, this parameter scaling approach makes the calculation
compatible with existing CUDA force kernels.
The NAMD implementation provides additional flexibility to
solute scaling by allowing different scaling factors for electrostatics,
van der Waals, and bonded interactions, as described in the
following section.
Solute scaling can be combined with replica exchange
to produce a powerful sampling enhancement method
that is highly transferable and provides higher efficiency
than traditional temperature exchange methods.
In the literature, this replica exchange solute scaling method
is known as REST2, due to its improvement of the earlier
REST (replica exchange solute tempering) method
that directly scaled the temperature of the solute.
Sample files are available in directory lib/replica/REST2,
with script file lib/replica/REST2/rest2_remd.namd
demonstrating use of solute scaling with multiple replicas.
The following parameters are used to control solute scaling:
- soluteScaling
 Is replica exchange solute tempering enabled?
Is replica exchange solute tempering enabled?
Acceptable Values: on or off
Default Value: off
Description: Specifies whether or not REST2 is enabled.
If set on, then soluteScaling must also be set.
- soluteScalingFactor
 Solute scaling factor
Solute scaling factor
Acceptable Values: non-negative
Default Value: 1.0
Description: This option sets the scaling factor 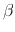 , and is typically set lower
than 1 to reduce potential energy barriers for the solute.
In a replica-exchange run, this option and the related options
soluteScalingFactorCharge and soluteScalingFactorVdw serve also as
Tcl scripting commands to update the corresponding parameters during a
simulation.
After using reinitatomsit is highly recommended that these
commands are repeated even if the corresponding parameters are not being
updated, to preserve their intended value.
, and is typically set lower
than 1 to reduce potential energy barriers for the solute.
In a replica-exchange run, this option and the related options
soluteScalingFactorCharge and soluteScalingFactorVdw serve also as
Tcl scripting commands to update the corresponding parameters during a
simulation.
After using reinitatomsit is highly recommended that these
commands are repeated even if the corresponding parameters are not being
updated, to preserve their intended value.
- soluteScalingFactorCharge
 Solute scaling factor for electrostatics
Solute scaling factor for electrostatics
Acceptable Values: non-negative
Default Value: soluteScalingFactor
Description: Scaling factor applied to just the electrostatics interactions.
If not specified, this is set to soluteScalingFactor.
- soluteScalingFactorVdw
 Solute scaling factor for van der Waals
Solute scaling factor for van der Waals
Acceptable Values: non-negative
Default Value: soluteScalingFactor
Description: Scaling factor applied to just the van der Waals interactions.
If not specified, this is set to soluteScalingFactor.
- soluteScalingFile
 PDB file with scaling flags
PDB file with scaling flags
Acceptable Values: UNIX filename
Default Value: coordinates
Description: PDB file used to flag solute atoms for scaling.
If undefined, this defaults to the coordinate PDB file.
- soluteScalingCol
 Column of PDB file
Column of PDB file
Acceptable Values: X, Y, Z, O, or B
Default Value: O
Description: Column of the PDB file used to flag solute atoms for scaling.
If undefined, this defaults to the O (occupancy) column.
A value of 1.0 marks the atom for scaling.
- soluteScalingAll
 Apply scaling also to bond and angle interactions?
Apply scaling also to bond and angle interactions?
Acceptable Values: on or off
Default Value: off
Description: If set on, scalingFactor is applied also to bond and angle interactions.
Otherwise, scalingFactor is applied only to dihedral, improper,
and crossterm interactions.




Next: Adaptive Tempering
Up: Accelerated Sampling Methods
Previous: Gaussian Accelerated Molecular Dynamics
Contents
Index
http://www.ks.uiuc.edu/Research/namd/
![]() ,
,