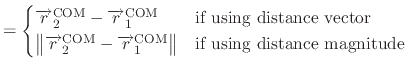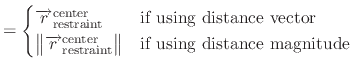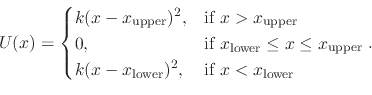



Next: Generalized Born Implicit Solvent
Up: Force Field Parameters
Previous: MARTINI Residue-Based Coarse-Grain Forcefield
Contents
Index
Subsections
Bond constraint parameters
The following describes the parameters for the position restraints feature of NAMD.
For historical reasons the term ``constraints'' has been carried over from X-PLOR.
This feature allows a restraining potential to each atom of an arbitrary set during the simulation.
- constraints
 are position restraints active?
are position restraints active? 
Acceptable Values: on or off
Default Value: off
Description: Specifies whether or not position restraints are active.
If it is set to off, then no position restraints are computed.
If it is set to on, the potential
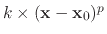 is applied to each atom.
Per-atom values for
is applied to each atom.
Per-atom values for 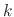 can be defined by either conskfile or conskcol, for
can be defined by either conskfile or conskcol, for
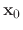 by consref, and for
by consref, and for 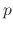 by consexp.
by consexp.
- consexp
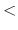 exponent for position restraint energy function
exponent for position restraint energy function 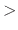
Acceptable Values: positive, even integer
Default Value: 2
Description: Exponent to be use in the position restraint energy function.
This value must be a positive integer, and only even values really make
sense. This parameter is used only if constraints is set to
on.
- consref
 PDB file containing restraint reference positions
PDB file containing restraint reference positions 
Acceptable Values: UNIX file name
Description: PDB file to use for reference positions for position restraints.
Each atom that has a positive force constant will be restrained about the position specified in this file.
- conskfile
 PDB file containing force constant values
PDB file containing force constant values 
Acceptable Values: UNIX filename
Description: PDB file to use for force constants for
position restraints.
- conskcol
 column of PDB file containing force constant
column of PDB file containing force constant 
Acceptable Values: X, Y, Z, O, or B
Description: Column of the PDB file to use for the position restraint force constant.
This parameter may specify any of the floating point fields of the PDB file,
either X, Y, Z, occupancy, or beta-coupling (temperature-coupling).
Regardless of which column is used, a value of 0 indicates that the atom
qshould not be restrained.
Otherwise, the value specified is used as the force constant for
that atom's restraining potential.
- constraintScaling
 scaling factor for position restraint energy function
scaling factor for position restraint energy function 
Acceptable Values: positive
Default Value: 1.0
Description: The position restraint energy function is multiplied by this parameter,
making it possible to gradually turn off restraints during equilibration.
This parameter is used only if constraints is set to
on.
- selectConstraints
 Restrain only selected Cartesian components of the coordinates?
Restrain only selected Cartesian components of the coordinates? 
Acceptable Values: on or off
Default Value: off
Description: This option is useful to restrain the positions of atoms to a plane or a line in space. If active,
this option will ensure that only selected Cartesian components of the coordinates are restrained.
E.g.: Restraining the positions of atoms to their current z values with no restraints
in x and y will allow the atoms to move in the x-y plane while retaining their original z-coordinate.
Restraining the x and y values will lead to free motion only along the z coordinate.
- selectConstrX
 Restrain X components of coordinates
Restrain X components of coordinates 
Acceptable Values: on or off
Default Value: off
Description: Restrain the Cartesian x components of the positions.
- selectConstrY
 Restrain Y components of coordinates
Restrain Y components of coordinates 
Acceptable Values: on or off
Default Value: off
Description: Restrain the Cartesian y components of the positions.
- selectConstrZ
 Restrain Z components of coordinates
Restrain Z components of coordinates 
Acceptable Values: on or off
Default Value: off
Description: Restrain the Cartesian z components of the positions.
The following describes the parameters for the group position restraints feature of NAMD.
Please note that this feature is only available in GPU-resident version of NAMD (NAMD3).
This feature allows restraining the center of mass (COM) of a group of atoms to a reference position
or another COM of group of atoms, during the simulation.
The restraining potential is defined as:
where 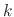 is restraint force constant defined by groupResK and
is restraint force constant defined by groupResK and
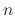 is the restraint exponent defined by groupResExp.
is the restraint exponent defined by groupResExp.
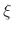 is the distance vector or length that connects the COM of
group 1 to the COM of group 2. In case of defining a reference position for
COM of group 1 (group1RefPos),
is the distance vector or length that connects the COM of
group 1 to the COM of group 2. In case of defining a reference position for
COM of group 1 (group1RefPos),
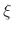 specifies the distance vector or length that
connects the reference position to the COM of group 2.
specifies the distance vector or length that
connects the reference position to the COM of group 2.
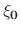 is the restraint center vector defined by groupResCenter.
is the restraint center vector defined by groupResCenter.
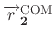 is calculated COM for the atoms in group 2,
where atom indices are defined in group2File or group2List.
is calculated COM for the atoms in group 2,
where atom indices are defined in group2File or group2List.
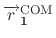 is either the reference position defined by
group1RefPos or calculated COM for the atoms in group 1,
where atom indices are defined in group1File or group1List.
is either the reference position defined by
group1RefPos or calculated COM for the atoms in group 1,
where atom indices are defined in group1File or group1List.
Once group position restraints are activated, NAMD outputs additional restraints information for
each restraint group. With the frequency of energy outputs, NAMD prints the following fields:
RES_TITLE: TS GROUP_NAME DISTANCE.X DISTANCE.Y DISTANCE.Z FORCE.X FORCE.Y FORCE.Z ENERGY
where "TS", "GROUP_NAME", "DISTANCE", "FORCE", "ENERGY" is timestep, assigned tag for the group restraint,
distance vector (Å)
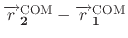 ,
applied restraint force (kcal/mol
,
applied restraint force (kcal/mol  Å) along
Å) along
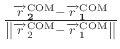 ,
and restraint energy (kcal/mol), respectively.
,
and restraint energy (kcal/mol), respectively.
Atoms may be held fixed during a simulation. NAMD avoids calculating most interactions in which all affected atoms are fixed unless fixedAtomsForces is specified.
Additional bond, angle, and dihedral energy terms may be applied to system,
allowing secondary or tertiary structure to be restrained, for example.
Extra bonded terms are not considered part of the molecular structure
and hence do not alter nonbonded exclusions.
The energies from extra bonded terms are included with the normal
bond, angle, and dihedral energies in NAMD output.
All extra bonded terms are harmonic potentials of the form
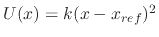 except dihedrals and impropers with
a non-zero periodicity specified, which use
except dihedrals and impropers with
a non-zero periodicity specified, which use
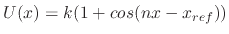 .
The only difference between dihedrals and
impropers is the output field that their potential energy is added to.
.
The only difference between dihedrals and
impropers is the output field that their potential energy is added to.
Due to a very old bug all NAMD releases prior to 2.13 have used the
MARTINI cosine-based angle potential function for all extra angles.
Since workflows may unknowingly depend on this undocumented behavior,
cosine-based angles remain the default, but a warning is printed
unless the desired behavior is specified via the new option
extraBondsCosAngles (defaults to ``on'', set to ``off'' to use
the normal harmonic angle potential function for all extra angles).
The extra bonded term implementation shares the parallel implementation
of regular bonded terms in NAMD, allowing large numbers of extra terms
to be specified with minimal impact on parallel scalability.
Extra bonded terms do not have to duplicate normal bonds/angles/dihedrals,
but each extra bond/angle/dihedral should only involve nearby atoms.
If the atoms involved are too far apart a bad global bond count will be
reported in parallel runs.
Extra bonded terms are enabled via the following options:
The extra bonds file(s) should contain lines of the following formats:
- bond <atom> <atom> <k> <ref>
- angle <atom> <atom> <atom> <k> <ref>
- dihedral <atom> <atom> <atom> <atom> <k> <ref>
- dihedral <atom> <atom> <atom> <atom> <k> <n> <ref>
- improper <atom> <atom> <atom> <atom> <k> <ref>
- improper <atom> <atom> <atom> <atom> <k> <n> <ref>
- wall <atom> <atom> <k> <lower> <upper>
- # <comment ...>
In all cases <atom> is a zero-based atom index
(the first atom has index 0),
<ref> is a reference distance in Å (bond) or angle in degrees (others),
and <k> is a spring constant in the potential energy function
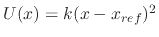 or, for dihedrals and impropers with
periodicity <n> specified and not 0,
or, for dihedrals and impropers with
periodicity <n> specified and not 0,
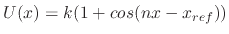 .
Note that
.
Note that 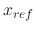 is only a minimum for the harmonic potential;
the sinusoidal potential has minima at
is only a minimum for the harmonic potential;
the sinusoidal potential has minima at
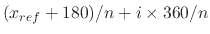 .
.
Use of wall implements a harmonic wall potential similar to
the Colvars harmonic wall restraint. The potential function is




Next: Generalized Born Implicit Solvent
Up: Force Field Parameters
Previous: MARTINI Residue-Based Coarse-Grain Forcefield
Contents
Index
http://www.ks.uiuc.edu/Research/namd/