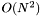 approach; for periodic systems it uses nearest image convention. This is intended only for very small systems or for testing.
approach; for periodic systems it uses nearest image convention. This is intended only for very small systems or for testing.
#include "mdapi/mdtypes.h"
#include "force/fparam.h"
#include "force/fdomain.h"
#include "force/fresult.h"
#include "force/fselect.h"
#include "force/defn.h"
#include "force/setup.h"
#include "force/compute.h"
Go to the source code of this file.
Defines | |
| #define | FORCE_FAIL (-1) |
Typedefs | |
| typedef Force_t | Force |
Functions | |
| Force * | force_create (ForceParam *fprm, ForceDomain *fdom, ForceSelect *fsel, const MD_Dvec initpos[]) |
| Constructor. | |
| void | force_destroy (Force *) |
| Destructor. | |
| int | force_compute (Force *, ForceResult *fres, const MD_Dvec pos[]) |
| Perform force evaluation. | |
| int | force_update_domain (Force *, ForceDomain *fdom) |
| Update the simulation cell domain. | |
| int | force_initialize (Force *, ForceParam *fprm, ForceDomain *fdom, ForceSelect *fsel, const MD_Dvec initpos[]) |
| Alternative constructor. | |
| void | force_cleanup (Force *) |
| Alternative destructor. | |
| const MD_Dvec * | force_get_poswrap (const Force *) |
| Return atomic coordinate offset array. | |
| const MD_Dvec * | force_get_scaled_coords (const Force *) |
| Return scaled atomic coordinate array. | |
| int32 ** | force_get_excl_list (const Force *) |
| Return exclusion lists. | |
| int32 ** | force_get_scaled14_list (const Force *) |
| Return scaled 1-4 exclusion lists. | |
| double | force_get_volume (const Force *) |
| const MD_Dvec * | force_get_cell_center (const Force *) |
| const MD_Dvec * | force_get_cell_vectors (const Force *) |
| const double * | force_get_cell_lengths (const Force *) |
| const MD_Dvec * | force_get_row_transform (const Force *) |
| int32 | force_get_cell_boundary (const Force *) |
| int32 | force_get_domain_update (const Force *) |
| int | force_setup_scaled_minmax (Force *) |
| const MD_Dvec * | force_get_scaled_minmax (const Force *) |
| int | force_setup_lattice (Force *, int32 k1, int32 k2, int32 k3) |
| const MD_Dvec * | force_get_lattice_spacings (const Force *) |
Force_t class evaluates the basic force field for biomolecules. Boundary conditions may be nonperiodic or periodic (or semi-periodic) cells. There are four helper container classes: ForceParam_t to define the force field parameters, ForceDomain_t to define the cell geometry, ForceResult_t which holds the results of the force and energy computation, and the optional class ForceSelect_t which can be used to evaluate some subset of bonds or to compute interaction energies between two disjoint subsets of atoms.The force evaluation can computes the following:
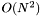 approach; for periodic systems it uses nearest image convention. This is intended only for very small systems or for testing.
approach; for periodic systems it uses nearest image convention. This is intended only for very small systems or for testing.Force_t class is used with four helper classes: ForceParam_t is configured at the beginning of the simulation to describe which aspects of the force field are to be evaluated, along with force field parameters and system topology data. ForceDomain_t defines the domain cell and boundary conditions. ForceResult_t provides containers for force and energy computation, providing buffer space for the accumulation of the different potential energies, atomic forces, and interaction energies. ForceSelect_t (optional) select for evaluation the energies from subsets of bonds or interaction energies and potentials between two disjoint subsets of atoms.ForceDomain_t cell vectors between calls to force_compute().The units are as outlined by the MDAPI documentation:
Caveats:
|
|
Return value from failed function call. |
|
|
Alternative destructor.
Use this to destroy a preallocated |
|
||||||||||||||||
|
Perform force evaluation.
The computed force and potential results are returned in the
|
|
||||||||||||||||||||
|
Constructor.
Force_t force evaluation object and initializes it. Must call this routine first. The user should not change the fprm after calling this routine. Any changes to fdom should be immediately followed by a call to force_update_domain(). If no selections desired, then fsel can be set to NULL; otherwise, no changes should be made to fsel. This routine does not compute the force, however, it is assumed that the initial atomic position coordinates initpos will not change more than the length of the ForceDomain_t cell vectors between calling this routine and all subsequent calls to force_compute().
|
|
|
Destructor.
Frees memory allocations made internally during setup and destroys the dynamically allocated |
|
|
Return exclusion lists.
This returns the exclusion lists created during |
|
|
Return atomic coordinate offset array.
This returns an array of position offsets that "wrap" the atomic coordinates used in the most recent call to |
|
|
Return scaled 1-4 exclusion lists.
The same format as returned by |
|
||||||||||||||||||||||||
|
Alternative constructor.
Use this to construct a preallocated
|
|
||||||||||||
|
Update the simulation cell domain.
ForceDomain_t fdom object provided to force_create(). Resets the transformation matrix and internal data storage needed for evaluation of nonbonded interactions.
|
 1.3.9.1
1.3.9.1