Dive into the world of computational biophysics through state-of-the-art techniques and tools in this comprehensive workshop. Our expert members from the NIH Center will guide you through a range of topics, including:

Molecular Dynamics Simulations
Learn the fundamentals and advanced applications using NAMD. Get to know about NAMD 3.0 and the new advancements in GPU-computing with the software developers.
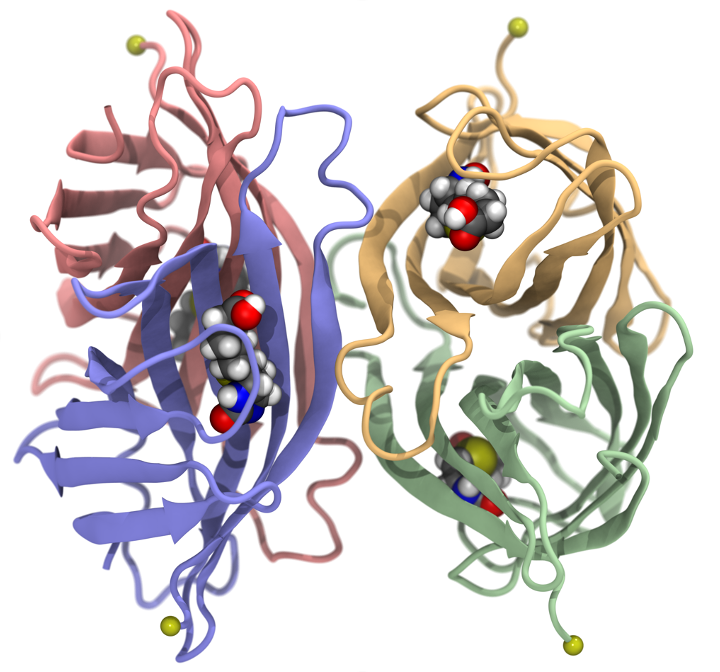
Biomolecular Visualization and Analysis
Gain hands-on experience with VMD. Have direct access to the software developers from UIUC, Auburn University and NVIDIA. You will be the first to try our new VMD 2.0, with completely modernized interface.
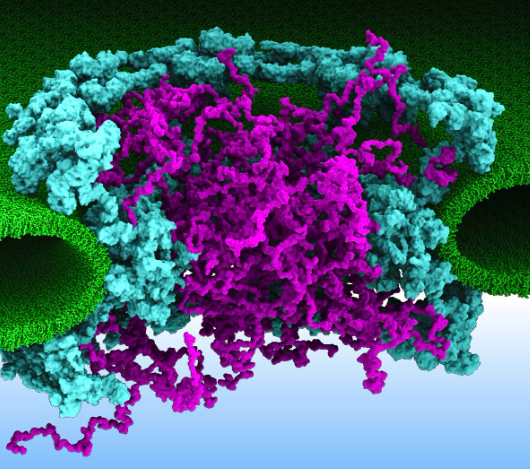
Nanotechnology Simulations
Explore new simulation techniques with ARBD. Learn how to simulate very large systems using Brownian Dynamics. Get to know the software developers and have a hands-on interaction with the software.










