


Next: Simulations of DNA permeation
Up: Bionanotechnology Tutorial
Previous: Introduction
Contents
Subsections
Simulation setup and protocols
In this unit you will learn to construct synthetic systems and simulate the passage of ions through a nanopore device.
In this section, we'll learn how to build a crystalline membrane from
its unit cell.
- 1
- Let's take a look at the unit cell in VMD. If you have not
already opened VMD, do so now. Open the Tk Console by
selecting Extensions
 Tk Console. Open the
directory with the files for this section and load the unit cell by
entering the following:
Tk Console. Open the
directory with the files for this section and load the unit cell by
entering the following:
| cd ``your working directory'' |
|
| cd bionano-tutorial-files |
|
| cd 1_build |
|
| mol new unit_cell_alpha.pdb |
|
Next, select Graphics  Representations....
In the Graphical Representations window, set the
Drawing Method to CPK. Now we can clearly see the
configuration of atoms in the unit cell. This configuration of eight
nitrogen atoms and six silicon atoms will form the basis for our
extended
Representations....
In the Graphical Representations window, set the
Drawing Method to CPK. Now we can clearly see the
configuration of atoms in the unit cell. This configuration of eight
nitrogen atoms and six silicon atoms will form the basis for our
extended
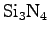 crystal.
crystal.
Figure 2:
Process of modeling a silicon nitride device. First the unit
cell is replicated to form a crystal membrane. This membrane is then
cut to a more convenient geometry. Finally, a pore is produced in the
membrane by the removal of atoms.
|
|
To generate a crystal membrane from the unit cell, we will
execute a script included with this tutorial. The main steps in the script
are as follows. First, we open an output PDB and write REMARK
lines specifying the geometry of the crystal. Next, we read the unit
cell PDB, extracting the records for each atom. We then generate the
crystal by repeatedly writing the atom records from the unit cell PDB
to the output PDB, albeit with new positions that are displaced by
periodic lattice vectors. In the following part of the tutorial, the
script, replicateCrystal.tcl, we will use to generate the
crystal is presented. Each portion of the script is preceded by text
describing how it works. If you'd like to move on with the tutorial
without examining the details of the script, simply skip ahead to
page ![[*]](crossref.gif) .
.
As will be common to most of the scripts presented in this tutorial,
the first section of the script defines variables that act as the
arguments of the script. The names of input and output files will
often appear here, as in the case below, where the name of the PDB
file containing the crystal's unit cell is stored in unitCellPdb and that of the resultant PDB is stored in outPdb.
The variables n1, n2, and n3 determine the number
of times that the unit cell will be replicated along the respective
crystal axis. The remaining variables describe the geometry of the
unit cell. The unit cell is a parallelepiped with sides of lengths
l1, l2, and l3 along directions given by the unit
vectors basisVector1, basisVector2, and basisVector3. Thus, the set of vectors {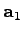 ,
,
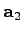 ,
, 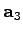 }, where
}, where
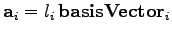 , generates the
translational symmetry of the lattice.
, generates the
translational symmetry of the lattice.
replicateCrystal.tcl
# Read the unit cell of a pdb and replicate n1 by n2 by n3 times.
# Input:
set unitCellPdb unit_cell_alpha.pdb
# Output:
set outPdb membrane.pdb
# Parameters:
# Choose n1 and n2 even if you wish to use cutHexagon.tcl.
set n1 6
set n2 6
set n3 6
set l1 7.595
set l2 7.595
set l3 2.902
set basisVector1 [list 1.0 0.0 0.0]
set basisVector2 [list 0.5 [expr sqrt(3.)/2.] 0.0]
set basisVector3 [list 0.0 0.0 1.0]
The two following Tcl procedures extract data from
the PDB. The first returns a list of 3D
vectors corresponding to the {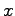
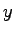
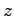 } coordinates of each atom
in the unit cell. The second simply extracts each line atom record
from the PDB and returns it as a list.
} coordinates of each atom
in the unit cell. The second simply extracts each line atom record
from the PDB and returns it as a list.
# Return a list with atom positions.
proc extractPdbCoords {pdbFile} {
set r {}
# Get the coordinates from the pdb file.
set in [open $pdbFile r]
foreach line [split [read $in] \n] {
if {[string equal [string range $line 0 3] "ATOM"]} {
set x [string trim [string range $line 30 37]]
set y [string trim [string range $line 38 45]]
set z [string trim [string range $line 46 53]]
lappend r [list $x $y $z]
}
}
close $in
return $r
}
# Extract all atom records from a pdb file.
proc extractPdbRecords {pdbFile} {
set in [open $pdbFile r]
set pdbLine {}
foreach line [split [read $in] \n] {
if {[string equal [string range $line 0 3] "ATOM"]} {
lappend pdbLine $line
}
}
close $in
return $pdbLine
}
Given the coordinates of all atoms in the unit cell, the displaceCell procedure shifts them by a lattice vector. In other
words, this procedure is where the crystal is actually
replicated--the basis on which the rest of the script rests.
# Shift a list of vectors by a lattice vector.
proc displaceCell {rUnitName i1 i2 i3 a1 a2 a3} {
upvar $rUnitName rUnit
# Compute the new lattice vector.
set rShift [vecadd [vecscale $i1 $a1] [vecscale $i2 $a2]]
set rShift [vecadd $rShift [vecscale $i3 $a3]]
set rRep {}
foreach r $rUnit {
lappend rRep [vecadd $r $rShift]
}
return $rRep
}
The procedure makePdbLine is essential to the correct formation of
the output PDB. The lines of the unit cell PDB obtained by extractPdbRecords are altered to reflect the new coordinates of the
translated unit cells.
# Construct a pdb line from a template line, index, resId, and coordinates.
proc makePdbLine {template index resId r} {
foreach {x y z} $r {break}
set record "ATOM "
set si [string range [format " %5i " $index] end-5 end]
set temp0 [string range $template 12 21]
set resId [string range " $resId" end-3 end]
set temp1 [string range $template 26 29]
set sx [string range [format " %8.3f" $x] end-7 end]
set sy [string range [format " %8.3f" $y] end-7 end]
set sz [string range [format " %8.3f" $z] end-7 end]
set tempEnd [string range $template 54 end]
# Construct the pdb line.
return "${record}${si}${temp0}${resId}${temp1}${sx}${sy}${sz}${tempEnd}"
}
The final procedure drives the script. The series of puts
commands near the top of the procedure store the geometry of the
crystal in REMARK lines in the output PDB. These lines will be
needed later when we modify the shape of the crystal. The lattice
vectors are defined by
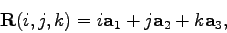
where 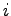 ,
, 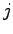 , and
, and 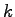 are integers. The main loop iterates through
all unique (
are integers. The main loop iterates through
all unique (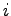 ,
,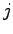 ,
,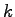 ) for
) for
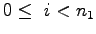 ,
,
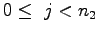 , and
, and
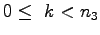 , producing a crystal.
, producing a crystal.
# Build the crystal.
proc main {} {
global unitCellPdb outPdb
global n1 n2 n3 l1 l2 l3 basisVector1 basisVector2 basisVector3
set out [open $outPdb w]
puts $out "REMARK Unit cell dimensions:"
puts $out "REMARK a1 $a1"
puts $out "REMARK a2 $a2"
puts $out "REMARK a3 $a3"
puts $out "REMARK Basis vectors:"
puts $out "REMARK basisVector1 $basisVector1"
puts $out "REMARK basisVector2 $basisVector2"
puts $out "REMARK basisVector3 $basisVector3"
puts $out "REMARK replicationCount $n1 $n2 $n3"
set a1 [vecscale $l1 $basisVector1]
set a2 [vecscale $l2 $basisVector2]
set a3 [vecscale $l3 $basisVector3]
set rUnit [extractPdbCoords $unitCellPdb]
set pdbLine [extractPdbRecords $unitCellPdb]
puts "\nReplicating unit $unitCellPdb cell $n1 by $n2 by $n3..."
# Replicate the unit cell.
set atom 1
set resId 1
for {set k 0} {$k < $n3} {incr k} {
for {set j 0} {$j < $n2} {incr j} {
for {set i 0} {$i < $n1} {incr i} {
set rRep [displaceCell rUnit $i $j $k $a1 $a2 $a3]
# Write each atom.
foreach r $rRep l $pdbLine {
puts $out [makePdbLine $l $atom $resId $r]
incr atom
}
incr resId
if {$resId > 9999} {
puts "Warning! Residue overflow."
set resId 1
}
}
}
}
puts $out "END"
close $out
puts "The file $outPdb was written successfully."
}
main
- 2
- To execute the script, enter
source replicateCrystal.tcl in the VMD Tk Console.
- 3
- We will now edit the script replicateCrystal.tcl in
order to make a thicker
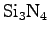 block. Open the file replicateCrystal.tcl in your text editor of choice, e.g., by typing
nedit replicateCrystal.tcl & in the terminal window. First,
change the line 8 to set outPdb block.pdb. Next change the
value of n3 by altering line 13 to read set n3 16. Save
the file and exit the text editor.
block. Open the file replicateCrystal.tcl in your text editor of choice, e.g., by typing
nedit replicateCrystal.tcl & in the terminal window. First,
change the line 8 to set outPdb block.pdb. Next change the
value of n3 by altering line 13 to read set n3 16. Save
the file and exit the text editor.
- 4
- To generate this thicker block of
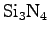 , execute the modified
script by entering source replicateCrystal.tcl as before.
, execute the modified
script by entering source replicateCrystal.tcl as before.
- 5
- We've now created two
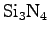 crystals. To view the first, type the following in the Tk Console window:
crystals. To view the first, type the following in the Tk Console window:
| mol delete all |
|
| mol new membrane.pdb |
|
This is the membrane that we will use for ionic current measurement and DNA translocation.
Notice that the cross section of the system in 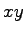 -plane is parallelogram.
-plane is parallelogram.
- 6
- Similarly, open the thicker block, which we'll use in Task 1.
Enter the following:
| mol delete all |
|
| mol new block.pdb |
|
Now we'll construct a nanopore in our
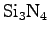 membranes.
membranes.
Our subsequent MD simulations will use periodic boundary conditions,
so the shape of our system must be such that the
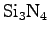 lattice matches
at the system's boundaries. A hexagonal prism shape can match this
lattice and is more convenient than the parallelpiped we just created
for housing a nanopore with a roughly circular cross section.
In the script for this
purpose, we first obtain the crystal geometry from the REMARK
lines in the PDB and write a file describing the hexagonal periodic
boundary conditions. Next, for convenience, we shift the crystal so
that its centroid coincides with the origin of the coordinate
system. We finally copy the atom records from the input PDB to the
output PDB, skipping those that do not lie within the hexagonal
prism. If you'd like to skip the details of this script, move on to
page
lattice matches
at the system's boundaries. A hexagonal prism shape can match this
lattice and is more convenient than the parallelpiped we just created
for housing a nanopore with a roughly circular cross section.
In the script for this
purpose, we first obtain the crystal geometry from the REMARK
lines in the PDB and write a file describing the hexagonal periodic
boundary conditions. Next, for convenience, we shift the crystal so
that its centroid coincides with the origin of the coordinate
system. We finally copy the atom records from the input PDB to the
output PDB, skipping those that do not lie within the hexagonal
prism. If you'd like to skip the details of this script, move on to
page ![[*]](crossref.gif) .
.
The first section again contains what serves as arguments to the
script. To save time when the script is altered to act on different
files, a file name prefix is defined which gives the output files
systematic names based on the name of the input file. In addition to
cutting the system to a hexagonal prism, the script cutHexagon.tcl also produces a boundary file (with a .bound
extension) that contains the periodic simulation cell vectors needed to form
bonds between atoms at the boundaries and run simulations in NAMD.
cutHexagon.tcl
# Remove atoms from a pdb outside of a hexagonal prism
# along the z-axis with a vertex along the x-axis.
# Also write a file with NAMD cellBasisVectors.
set fileNamePrefix membrane
# Input:
set pdbIn ${fileNamePrefix}.pdb
# Output:
set pdbOut ${fileNamePrefix}_hex.pdb
set boundaryFile ${fileNamePrefix}_hex.bound
set pdbTemp tmp.pdb
This procedure executes VMD's measure center method to center
the system at the origin, which is done for convenience.
# Write a pdb with the system centered.
proc centerPdb {pdbIn pdbOut} {
mol new $pdbIn
set all [atomselect top all]
set cen [measure center $all]
$all moveby [vecinvert $cen]
$all writepdb $pdbOut
$all delete
mol delete top
}
The procedure readGeometry extracts the crystal geometry from
the REMARK lines we added to the PDB in last script and writes
the boundary file mentioned above.
# Read the geometry of the system and write the boundary file.
# Return the radius of the hexagon.
proc readGeometry {pdbFile boundaryFile} {
# Extract the remark lines from the pdb.
mol new $pdbFile
set remarkLines [lindex [molinfo top get remarks] 0]
foreach line [split $remarkLines \n] {
if {![string equal [string range $line 0 5] "REMARK"]} {continue}
set tok [concat [string range $line 7 end]]
set attr [lindex $tok 0]
set val [lrange $tok 1 end]
set remark($attr) $val
puts "$attr = $val"
}
mol delete top
# Deterimine the lattice vectors.
set vector1 [vecscale $remark(basisVector1) $remark(a1)]
set vector2 [vecscale $remark(basisVector2) $remark(a2)]
set vector3 [vecscale $remark(basisVector3) $remark(a3)]
foreach {n1 n2 n3} $remark(replicationCount) {break}
set pbcVector1 [vecadd [vecscale $vector1 [expr $n1/2]] \
[vecscale $vector2 [expr $n2/2]]]
set pbcVector2 [vecadd [vecscale $vector1 [expr -$n1/2] ] \
[vecscale $vector2 [expr $n2]]]
set pbcVector3 [vecscale $vector3 $n3]
puts ""
puts "PERIODIC VECTORS FOR NAMD:"
puts "cellBasisVector1 $pbcVector1"
puts "cellBasisVector2 $pbcVector2"
puts "cellBasisVector3 $pbcVector3"
puts ""
set radius [expr 2.*[lindex $pbcVector1 0]/3.]
puts "The radius of the hexagon: $radius"
# Write the boundary condition file.
set out [open $boundaryFile w]
puts $out "radius $radius"
puts $out "cellBasisVector1 $pbcVector1"
puts $out "cellBasisVector2 $pbcVector2"
puts $out "cellBasisVector3 $pbcVector3"
close $out
return $radius
}
Here, in the procedure cutHexagon, we read each atom record
from the input PDB and extract the serial number and coordinates. The
record is then written to the output PDB if and only if the position
of the atom is within a hexagon of radius 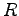 in the
in the 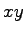 -plane,
centered at the origin, which has a vertex along the
-plane,
centered at the origin, which has a vertex along the 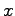 -axis. All
three of the following geometric criteria must hold:
-axis. All
three of the following geometric criteria must hold:
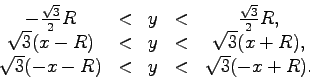
proc cutHexagon {r pdbIn pdbOut} {
set sqrt3 [expr sqrt(3.0)]
# Open the pdb to extract the atom records.
set out [open $pdbOut w]
set in [open $pdbIn r]
set atom 1
foreach line [split [read $in] \n] {
set string0 [string range $line 0 3]
# Just write any line that isn't an atom record.
if {![string match $string0 "ATOM"]} {
puts $out $line
continue
}
# Extract the relevant pdb fields.
set serial [string range $line 6 10]
set x [string range $line 30 37]
set y [string range $line 38 45]
set z [string range $line 46 53]
# Check the hexagon bounds.
set inHor [expr abs($y) < 0.5*$sqrt3*$r]
set inPos [expr $y < $sqrt3*($x+$r) && $y > $sqrt3*($x-$r)]
set inNeg [expr $y < $sqrt3*($r-$x) && $y > $sqrt3*(-$x-$r)]
# If atom is within the hexagon, write it to the output pdb
if {$inHor && $inPos && $inNeg} {
# Make the atom serial number accurate if necessary.
if {[string is integer [string trim $serial]]} {
puts -nonewline $out "ATOM "
puts -nonewline $out \
[string range [format " %5i " $atom] end-5 end]
puts $out [string range $line 12 end]
} else {
puts $out $line
}
incr atom
}
}
close $in
close $out
}
In the main part of the script, we extract the radius of the hexagon
and write the boundary file, center the crystal, and finally cut
the crystal into a hexagonal prism.
set radius [readGeometry $pdbIn $boundaryFile]
centerPdb $pdbIn $pdbTemp
cutHexagon $radius $pdbTemp $pdbOut
- 1
- Enter source cutHexagon.tcl in the Tk Console.
The script acts on membrane.pdb, producing the file membrane_hex.pdb.
We also need to cut block.pdb to a hexagonal prism.
- 2
- Open cutHexagon.tcl in your text editor. Change line 7
to read set fileNamePrefix block and save the file. Execute the
script by reentering
source cutHexagon.tcl in the
Tk Console.
- 3
- Let's look at our system in VMD to make sure it has been cut
into a hexagonal prism correctly. Type:
| mol delete all |
|
| mol load pdb membrane_hex.pdb |
|
- 4
- Also, look at the second system. Enter mol delete all
and mol load pdb block_hex.pdb in the Tk Console.
Now we'll shape our crystals into nanopore devices. The script drillPore.tcl has been designed for this purpose. We'll produce a
pore with the shape of two intersecting cones, which has
hourglass-like cross sections in the 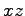 - or
- or 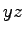 - planes. First, we
read the length of the pore along the
- planes. First, we
read the length of the pore along the 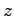 -axis from the boundary
file. Subsequently, we remove atoms from the PDB file that are within the
pore. You can skip the details of this script by turning to
page
-axis from the boundary
file. Subsequently, we remove atoms from the PDB file that are within the
pore. You can skip the details of this script by turning to
page ![[*]](crossref.gif) .
.
The parameters radiusMin and radiusMax define
the minimum and maximum radius of the double-cone pore.
drillPore.tcl
# Cut a double-cone pore in a membrane.
# Parameters:
set radiusMin 8
set radiusMax 15
# Input:
set pdbIn membrane_hex.pdb
set boundaryFile membrane_hex.bound
# Output:
set pdbOut pore.pdb
set boundaryOut pore.bound
This procedure extracts the length of the pore along the 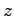 -axis,
which is necessary for defining the geometry of the double cone pore.
-axis,
which is necessary for defining the geometry of the double cone pore.
# Get cellBasisVector3_z from the boundary file.
proc readLz {boundaryFile} {
set in [open $boundaryFile r]
foreach line [split [read $in] \n] {
if {[string match "cellBasisVector3 *" $line]} {
set lz [lindex $line 3]
break
}
}
close $in
return $lz
}
In a membrane of thickness 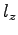 , the cylindrical coordinate
, the cylindrical coordinate 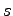 that
corresponds to the radius of the pore at height
that
corresponds to the radius of the pore at height 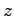 for a double cone
with a center radius of
for a double cone
with a center radius of
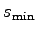 and a maximum radius of
and a maximum radius of
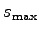 is given by
is given by
Whether the point 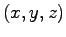 is within the pore is
determined by
is within the pore is
determined by
Later, in Task 1, you will modify a similar
procedure to produce a topologically more complicated pore.
# Determine whether the position {x y z} is inside the pore and
# should be deleted.
proc insidePore {x y z sMin sMax} {
# Get the radius for the double cone at this z-value.
set s [expr $sMin + 2.0*($sMax-$sMin)/$lz*abs($z)]
return [expr $x*$x + $y*$y < $s*$s]
}
The final procedure is nearly identical to the cutHexagon
procedure in cutHexagon.tcl. It writes only lines satisfying
geometrical constraints, this time given by the result of the
procedure insidePore.
proc drillPore {sMin sMax lz pdbIn pdbOut} {
set sqrt3 [expr sqrt(3.0)]
# Open the pdb to extract the atom records.
set out [open $pdbOut w]
set in [open $pdbIn r]
set atom 1
foreach line [split [read $in] \n] {
set string0 [string range $line 0 3]
# Just write any line that isn't an atom record.
if {![string match $string0 "ATOM"]} {
puts $out $line
continue
}
# Extract the relevant pdb fields.
set serial [string range $line 6 10]
set x [string range $line 30 37]
set y [string range $line 38 45]
set z [string range $line 46 53]
# If atom is outside the pore, write it to the output pdb.
# Otherwise, exclude it from the resultant pdb.
if {![insidePore $x $y $z $sMin $sMax]} {
# Make the atom serial number accurate if necessary.
if {[string is integer [string trim $serial]]} {
puts -nonewline $out "ATOM "
puts -nonewline $out \
[string range [format " %5i " $atom] end-5 end]
puts $out [string range $line 12 end]
} else {
puts $out $line
}
incr atom
}
}
close $in
close $out
}
set lz [readLz $boundaryFile]
drillPore $radiusMin $radiusMax $lz $pdbIn $pdbOut
- 5
- In the Tk Console, enter source drillPore.tcl.
- 6
- Let's examine the pore we just created in VMD. Enter mol
delete all and mol load pdb pore.pdb in the Tk
Console. Setting the Drawing Method to VDW and the
Selected Atoms edit box to abs(y) < 5 should make the
double pore cross section apparent.
- 7
- The file produced, pore.pdb, needs an accompanying
boundary file. In the Tk Console, enter cp membrane_hex.bound pore.bound.
We've constructed two crystalline membranes and, from them,
two nanopores; however, we have only generated atom coordinates. We
have not defined bonds of any sort between the atoms. In this section,
we'll construct a PSF file that describes the bonds (connections
between two atoms) and angles (connections between three
atoms) in our systems as well as other items needed for subsequent MD
simulations. To do this we'll use the script siliconNitridePsf.tcl. This script is somewhat longer than those we
have seen thus far, so its description has been left to the appendix.
A quick synopsis of the script's operation is as follows. The first
step is to find bonds simply by searching for atoms that are within
some threshold distance of one another. However, this step misses
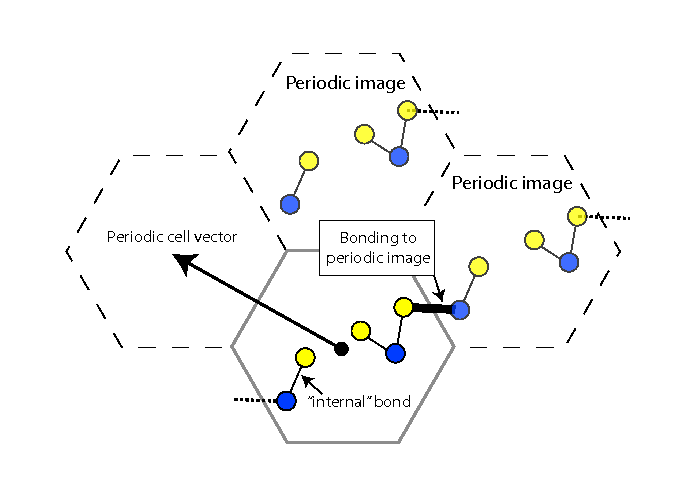
bonds that exist across the periodic boundaries. To find these, we
displace the system by the periodic cell vectors and find bonds
between the original system and its periodic image
(Fig. 3). Next we determine the angles and then
finally write all of the information to a PSF file.
Figure 3:
Bonding to periodic images. The periodic image is produced by
translating the system by a periodic cell vector. To find bonds across
the periodic boundary, a distance search is performed between the
original coordinates of the atoms and those in each periodic image.
|
|
You may be used to calling upon psfgen to produce the structure
files for proteins and other biomolecules. This would be possible for
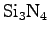 as well. However, due to the nature of the material, it is
somewhat more straightforward to generate the PSF directly as we do
with this script.
as well. However, due to the nature of the material, it is
somewhat more straightforward to generate the PSF directly as we do
with this script.
- 1
- We'll now build the PSF structure file for our membrane. Type
source siliconNitridePsf.tcl in the Tk Console. The
structure information for our pore is now contained in pore.psf.
- 2
- Let's also build the structure for the pristine
membrane. Change line 5 of siliconNitridePsf.tcl to set
fileNamePrefix membrane_hex. Since we will use the pristine membrane
to calibrate the dielectric constant of the silicon nitride, we
do not want any surfaces. Change line 16 to read set zPeriodic
1. Save the script and then execute it.
- 3
- Take a look at the system in VMD by entering mol delete
all and mol load psf pore.psf pdb pore.pdb in the Tk
Console. Select Graphics
 Representations.... Notice that there appear to be bonds
crisscrossing the pore. This occurs because VMD can't correctly
display bonds across the periodic boundaries. Set the drawing method
Drawing Method to VDW, which does not illustrate
bonds. The pore should now be clearly visible.
Representations.... Notice that there appear to be bonds
crisscrossing the pore. This occurs because VMD can't correctly
display bonds across the periodic boundaries. Set the drawing method
Drawing Method to VDW, which does not illustrate
bonds. The pore should now be clearly visible.
Bionanotechnology enters uncharted territory by placing
together biomolecules and synthetic materials that have rarely been
studied in contact. In addition, simulations of inorganic solids such
as
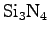 usually employ vastly different methods than those used in
computational molecular biology. Thus, simulating systems with both
synthetic and biomolecular constituents is challenging and, in
general, an unsolved problem. Because much research in
bionanotechnology involves electrostatic interactions between
biomolecules and silicon-based materials, we'll focus on getting our
usually employ vastly different methods than those used in
computational molecular biology. Thus, simulating systems with both
synthetic and biomolecular constituents is challenging and, in
general, an unsolved problem. Because much research in
bionanotechnology involves electrostatic interactions between
biomolecules and silicon-based materials, we'll focus on getting our
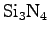 model to reproduce experimental data for just one property: the
dielectric constant. With this model we can expect to have a realistic
electric field within the pore.
model to reproduce experimental data for just one property: the
dielectric constant. With this model we can expect to have a realistic
electric field within the pore.
To determine the dielectric constant, we will apply an electric field
to a block of
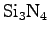 with no free surfaces and measure the electric
dipole moment. Hence, we will use the structure membrane_hex.psf
that we generated in the last section, for which we generated bonds
along all three lattice directions.
with no free surfaces and measure the electric
dipole moment. Hence, we will use the structure membrane_hex.psf
that we generated in the last section, for which we generated bonds
along all three lattice directions.
- 1
- Type cd ../2_calibrate/ in the Tk Console.
- 2
- Open the
parameter file par_silicon_ions_NEW0.1.inp in your text
editor. Notice that the file has three sections. The first two give energy
function parameters for harmonic bonds and harmonic angle bending
between two bonds, respectively. The last gives the parameters for the
non-bonded interactions. You may close the file now.
We take the non-bonded parameters, as well as the values for the
partial charges on the
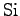 and
and
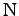 atoms in
siliconNitridePsf.tcl, from quantum mechanical calculations
using the biased Hessian method (John A. Wendell and William
A. Goddard III, Journal of Chemical Physics 97,
5048-5062 (1992)). However, the bonded interactions from the same
source lead to a dielectric constant that is practically the same as a
vacuum (1.0). To overcome this, we set the bonded interaction
constants to be much lower than those given in the reference. In this
section, we'll set them to 0.1
atoms in
siliconNitridePsf.tcl, from quantum mechanical calculations
using the biased Hessian method (John A. Wendell and William
A. Goddard III, Journal of Chemical Physics 97,
5048-5062 (1992)). However, the bonded interactions from the same
source lead to a dielectric constant that is practically the same as a
vacuum (1.0). To overcome this, we set the bonded interaction
constants to be much lower than those given in the reference. In this
section, we'll set them to 0.1
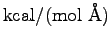 . To match
the experimental dielectric constant we include in our force field
harmonic restraints, which can easily be applied in NAMD, that pull
each atom of
. To match
the experimental dielectric constant we include in our force field
harmonic restraints, which can easily be applied in NAMD, that pull
each atom of
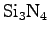 towards its equilibrium position in the
towards its equilibrium position in the
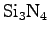 crystal. It is the spring constant associated with these constraint
forces that we will calibrate to reproduce the
experimentally-determined dielectric constant of
crystal. It is the spring constant associated with these constraint
forces that we will calibrate to reproduce the
experimentally-determined dielectric constant of
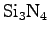 .
.
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\end{minipage} \begin{min...
...e {\tt chargeSi} in the PSF
generating script (See Appendix).}
\end{minipage} }](img37.gif)
- 3
- The Tcl script constrainSilicon.tcl produces PDB files
where the spring constant is placed in the B (known in VMD as beta)
column of the PDB. Open the script in your text editor. A constraint
PDB will be produced for each spring constant (
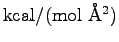 ) in the list defined in line 7. We'll determine the
dielectric constant for values 1.0 and 10.0. Hence, change line 7 of
the script to set betaList {1.0 10.0}. Execute constrainSilicon.tcl, whose contents follow.
) in the list defined in line 7. We'll determine the
dielectric constant for values 1.0 and 10.0. Hence, change line 7 of
the script to set betaList {1.0 10.0}. Execute constrainSilicon.tcl, whose contents follow.
constrainSilicon.tcl
# Add harmonic constraints to silicon nitride.
# Parameters:
# Spring constant in kcal/(mol A^2)
set betaList {1.0}
set selText "resname SIN"
set surfText "(name \"SI.*\" and numbonds<=3) \
or (name \"N.*\" and numbonds<=2)"
# Input:
set psf ../1_build/membrane_hex.psf
set pdb ../1_build/membrane_hex.pdb
# Output:
set restFilePrefix siliconRest
mol load psf $psf pdb $pdb
set selAll [atomselect top all]
# Set the spring constants to zero for all atoms.
$selAll set occupancy 0.0
$selAll set beta 0.0
# Select the silicon nitride.
set selSiN [atomselect top $selText]
# Select the surface.
set selSurf [atomselect top "(${selText}) and (${surfText})"]
foreach beta $betaList {
# Set the spring constant for SiN to this beta value.
$selSiN set beta $beta
# Constrain the surface 10 times more than the bulk.
$selSurf set beta [expr 10.0*$beta]
# Write the constraint file.
$selAll writepdb ${restFilePrefix}_${beta}.pdb
}
$selSiN delete
$selSurf delete
$selAll delete
mol delete top
- 4
- Since the silicon atoms are already in their equilibrium positions,
we'll forgo the energy minimization step in the usual simulation sequence.
Instead, we'll start by raising the temperature gradually to 295 K.
During this time, we'll use constraints of 1.0
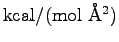 .
.
Before we start, however, we need to put the system dimensions in the
NAMD configuration file eq1.namd. Open it and 1_build/membrane_hex.bound (if you did not use InorganicBuilder),
which we generated in Section 1.2, in your text editor. If you used
InorganicBuilder, refer instead to the vectors you recorded. Copy the
values of cellBasisVector1, cellBasisVector2, and cellBasisVector3 into lines 8, 9, and 10, respectively, of the
configuration file. Also, examine the constraint parameters at the
bottom of the file. Save the configuration file and exit the text
editor.
- 5
- Enter namd2 eq1.namd > eq1.log to raise the system's temperature. This may take a couple of minutes.
- 6
- To equilibrate the system at constant temperature, enter namd2 eq2.namd > eq2.log.
- 7
- Next we compute the dielectric constant for each constraint
value. To do this, we calculate the difference between the dipole
moments of identical systems with and without an applied electric field.
Open the files field.namd
and null.namd in your text editor. Modify line 2 to read
set constraint 1.0. First simulate the system without the
applied electric field by entering namd2 null.namd >!
null1.0.log and then with a field of 16 kcal/(mol Å e) by entering
namd2 field.namd >! field1.0.log. Do the same for the other
constraint value, i.e., alter the variable constraint in field.namd and null.namd and run NAMD.
- 8
- We'll now compute the electric dipole moment for each run and
from these calculate the dielectric constant for the material. Open
the script dipoleMomentZDiff.tcl in your text editor. The
script operates by loading DCD trajectory files for the system with
and without an applied field. We then compute the dipole moment for
each frame and write the time (ns) in the first column and the
difference in the dipoles (e Å) in the second column of a text
file.
The values of dcdFreq and timestep, taken from the NAMD
configuration file, allow us to determine the time between the frames
of the DCD trajectory file. We'll set the variable startFrame
to 4 to give the system 500 fs to equilbrate before computing the
dipole moment. The electric dipole moment is computed by VMD's measure dipole command which employs the following formula. For a set
of 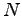 atoms with partial charges
atoms with partial charges
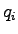 and positions
and positions 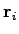 the electric dipole moment is
the electric dipole moment is
where
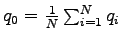 . Subtraction of
. Subtraction of 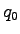 , the
monopole component, makes the result independent of the choice of the
origin. Finally, the script computes the average of the difference in
the dipole moments and the associated standard error.
, the
monopole component, makes the result independent of the choice of the
origin. Finally, the script computes the average of the difference in
the dipole moments and the associated standard error.
dipoleMomentZDiff.tcl
# Calculate dipole moment of the selection
# for a trajectory.
set constraint 10.0
set dcdFreq 100
set selText "all"
set startFrame 0
set timestep 1.0
# Input:
set psf ../1_build/membrane_hex.psf
set dcd field${constraint}.dcd
set dcd0 null${constraint}.dcd
# Output:
set outFile dipole${constraint}.dat
# Get the time change between frames in femtoseconds.
set dt [expr $timestep*$dcdFreq]
# Load the system.
set traj [mol load psf $psf dcd $dcd]
set sel [atomselect $traj $selText]
set traj0 [mol load psf $psf dcd $dcd0]
set sel0 [atomselect $traj0 $selText]
# Choose nFrames to be the smaller of the two.
set nFrames [molinfo $traj get numframes]
set nFrames0 [molinfo $traj0 get numframes]
if {$nFrames0 < $nFrames} {set nFrames $nFrames}
puts [format "Reading %i frames." $nFrames]
# Open the output file.
set out [open $outFile w]
# Start at "startFrame" and move forward, computing
# the dipole moment at each step.
set sum 0.
set sumSq 0.
set n 1
puts "t (ns)\tp_z (e A)\tp0_z (e A)\tp_z-p0_z (e A)"
for {set f $startFrame} {$f < $nFrames && $n > 0} {incr f} {
$sel frame $f
$sel0 frame $f
# Obtain the dipole moment along z.
set p [measure dipole $sel]
set p0 [measure dipole $sel0]
set z [expr [lindex $p 2] - [lindex $p0 2]]
# Get the time in nanoseconds for this frame.
set t [expr ($f+0.5)*$dt*1.e-6]
puts $out "$t $z"
puts -nonewline [format "FRAME %i: " $f]
puts "$t\t[lindex $p 2]\t[lindex $p0 2]\t$z"
set sum [expr $sum + $z]
set sumSq [expr $sumSq + $z*$z]
}
close $out
# Compute the mean and standard error.
set mean [expr $sum/$nFrames]
set meanSq [expr $sumSq/$nFrames]
set se [expr sqrt(($meanSq - $mean*$mean)/$nFrames)]
puts ""
puts "********Results: "
puts "mean dipole: $mean"
puts "standard error: $se"
mol delete top
mol delete top
- 9
- Execute the script dipoleMomentZDiff.tcl twice, setting
constraint (in line 6 of the script) to each of the values in our simulations. Be sure to write down the mean dipole and standard error for each.
- 10
- Plot the time versus dipole moment data stored the resulting
files dipole10.0.dat and dipole1.0.dat. You should see
that the dipole moments are changing little with time by the end of
the simulation and that the values are significantly different for the
two different constraint parameters.
- 11
- To calculate the dielectric constant we apply the formula
where 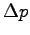 is the magnitude of the difference in the dipole
moment between identical systems with and without an electric field,
is the magnitude of the difference in the dipole
moment between identical systems with and without an electric field,
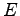 is the magnitude of the applied electric field, and
is the magnitude of the applied electric field, and 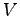 is the
volume of the system dielectric material (See Dong Xu, et al.,
The Journal of Physical Chemistry 100, 12108-12121
(1996) for further discussion). The permittivity of free space is given
in NAMD units by
is the
volume of the system dielectric material (See Dong Xu, et al.,
The Journal of Physical Chemistry 100, 12108-12121
(1996) for further discussion). The permittivity of free space is given
in NAMD units by
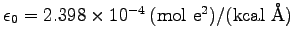 . We can calculate the volume of our
hexagonal prism by
. We can calculate the volume of our
hexagonal prism by
where 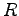 is the radius of the hexagon and
is the radius of the hexagon and 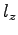 is the height of the
prism. Obtaining
is the height of the
prism. Obtaining 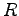 and
and 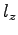 from membrane_hex.bound, we
find
from membrane_hex.bound, we
find
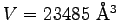 . Given that
. Given that
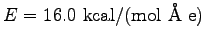 calculate the dielectric constants
for the two constraint values using the mean dipole values. Note that
you can use the form Tk Console as a calculator by typing expr commands. Is the difference in the dielectric constant between
the two significant?
calculate the dielectric constants
for the two constraint values using the mean dipole values. Note that
you can use the form Tk Console as a calculator by typing expr commands. Is the difference in the dielectric constant between
the two significant?
This section is only meant to be a demonstration of how the
calibration is performed. Sampling the entire parameter space takes a
good deal of time, but you should now have a good understanding of how
to calibrate the constraints to reproduce the experimental dielectric
constant. In subsequent sections, we will use a parameter file with
the bond constants set to 5.0
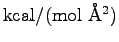 and a
constraint file with constants of 1.0
and a
constraint file with constants of 1.0
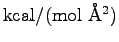 ,
which have been found to be optimal by the procedure above.
,
which have been found to be optimal by the procedure above.
Now that we've demonstrated how to calibrate the force field of
our
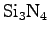 model, we're ready to prepare our nanopore for
simulations.
model, we're ready to prepare our nanopore for
simulations.
- 1
- In the Tk Console, type cd
../3_solvate/.
All biological systems rely on water to function. If our synthetic
device is to interact with them, it must be immersed in water.
- 2
- Open the system we wish to solvate by entering mol load psf ../1_build/pore.psf pdb ../1_build/pore.pdb in the Tk Console.
- 3
- To open the Solvate plugin, select Extensions
 Modeling
Modeling  Add Solvation Box from the VMD menu.
Add Solvation Box from the VMD menu.
- 4
- You should already see ../1_build/pore.psf and ../1_build/pore.pdb in the edit boxes labeled PSF and PDB, respectively. Set Output to pore_solv. Since we
wish to have water above and below the membrane, set the minimum and
maximum Box Padding in the direction z to 20.
- 5
- Press Solvate.
- 6
- Notice that the Solvate plugin adds the water in a right
rectangular prism, which does not conform to our hexagonal prism
periodic boundary conditions. Type mol delete all in the
Tk Console.
We'll now remove water from outside of the hexagonal boundaries with
the script cutWaterHex.tcl. It uses VMD's atom selection
interface to obtain the set {segname, resid, name}, which uniquely specifies each atom, for all atoms violating
the geometric constraints that we used in Section 1.2 to cut a hexagon
from our crystal. Then by applying the psfgen command delatom, violating atoms are deleted. We estimate the radius
of the hexagon with the measure minmax command provided by
VMD.
cutWaterHex.tcl
# This script will remove water from psf and pdf outside of a
# hexagonal prism along the z-axis.
package require psfgen 1.3
# Input:
set psf pore_solv.psf
set pdb pore_solv.pdb
# Output:
set psfFinal pore_hex.psf
set pdbFinal pore_hex.pdb
# Parameters:
# The radius of the water hexagon is reduced by "radiusMargin"
# from the pore hexagon. The distance is in angstroms.
set radiusMargin 0.5
# This is the stuff that is removed.
set waterText "water or ions"
# This selection forms the basis for the hexagon.
set selText "resname SIN"
# Load the molecule.
mol load psf $psf pdb $pdb
# Find the system dimensions.
set sel [atomselect top $selText]
set minmax [measure minmax $sel]
$sel delete
set size [vecsub [lindex $minmax 1] [lindex $minmax 0]]
foreach {size_x size_y size_z} $size {break}
# This is the hexagon's radius.
if {[expr $size_x > $size_y]} {
set rad [expr 0.5*$size_x]
} else {
set rad [expr 0.5*$size_y]
}
set r [expr $rad - $radiusMargin]
# Find water outside of the hexagon.
set sqrt3 [expr sqrt(3.0)]
# Check the middle rectangle.
set check "($waterText) and ((abs(x) < 0.5*$r and abs(y) > 0.5*$sqrt3*$r) or"
# Check the lines forming the nonhorizontal sides.
set check [concat $check "(y > $sqrt3*(x+$r) or y < $sqrt3*(x-$r) or"]
set check [concat $check "y > $sqrt3*($r-x) or y < $sqrt3*(-x-$r)))"]
set w [atomselect top $check]
set violators [lsort -unique [$w get {segname resid}]]
$w delete
# Remove the offending water molecules.
puts "Deleting the offending water molecules..."
resetpsf
readpsf $psf
coordpdb $pdb
foreach waterMol $violators {
delatom [lindex $waterMol 0] [lindex $waterMol 1]
}
writepsf $psfFinal
writepdb $pdbFinal
mol delete top
- 7
- Enter source cutWaterHex.tcl to
remove water outside of the hexagonal boundaries.
- 8
- Open the new structure by entering mol load psf
pore_hex.psf pdb pore_hex.pdb. Does the system now conform to a
hexagonal prism?
Many biomolecules are sensitive to the ionic strength of the
surrounding solvent; therefore, salt is added to the solutions used in
experiments to mimic physiological conditions. In addition, ions
facilitate measurements of small currents in nanopore systems by
substantially increasing the conductivity of the solution.
- 9
- To open the Autoionize plugin, select Extensions
 Modeling
Modeling  Add Ions from the VMD menu.
Add Ions from the VMD menu.
- 10
- You should already see pore_hex.psf and pore_hex.pdb in the edit boxes labeled PSF and PDB,
respectively. Set Output to pore_all. Since we wish
to have a 2 mol/kg KCl concentration, set Concentration to 4.
Set both Min. distance from molecule and Min. distance
between ions to 2. Also, because we are using a KCl solution instead
of NaCl, select the checkbox labeled Switch to KCl instead of
NaCl.
- 11
- Execute the Autoionize plugin by pressing Autoionize.
- 12
- For convenience, copy the solvated structure into the directory
for the next section by typing cp pore_all.psf ../4_current/
and cp pore_all.pdb ../4_current/.
In experiment, ionic current is a macroscopic quantity that gives
insight into nanoscale processes. Ionic current measurements are used
to characterize single nanopores and their interactions with
biological molecules. In this subsection, we'll learn to simulate our
nanopore system with an applied voltage and calculate the ionic
current from the trajectory.
- 1
- Enter cd ../4_current/ in the Tk Console to
change to the directory for this subsection. Be sure that you have
copied the files pore_all.psf and pore_all.pdb into this
directory as instructed at the end of the last subsection.
- 2
- First we need to generate the constraint file using the
parameters that reproduce the experimental dielectric constant. In the
Tk Console, enter
source
constrainSilicon.tcl.
- 3
- Now we need to equilibrate our system. We'll start by
performing energy minimization. Take a look at the NAMD configuration
file eq0.namd in your text editor. The values given for cellBasisVector1 and cellBasisVector2 match those given in
../1_build/membrane_hex.bound. If you used InorganicBuilder
to generate the pore, you should replace values with your own. The
third basis vector is dependent on the size of the water box we added.
To determine it, in the Tk Console window (Extensions
 Tk Console) type the following commands:
Tk Console) type the following commands:
| mol delete all |
|
| mol load psf
pore_all.psf pdb pore_all.pdb |
|
| set all [atomselect top
all] |
|
| set minmax [measure minmax $all] |
|
| set lz
[expr [lindex $minmax 1 2]-[lindex $minmax 0 2]] |
|
| $all
delete |
|
The value of lz gives us the size (Å) of the system along the
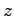 -axis. We don't want to put this value directly into the NAMD
configuration file, however.
-axis. We don't want to put this value directly into the NAMD
configuration file, however.
It is better for a few water molecules to be crowded at the ends at
this point than risk introducing a vacuum region at the ends. While
the minimization step can easily rearrange water molecules that have
been placed too close together due to wrapping at the periodic
boundaries, small regions of vacuum can cause inaccuracies in
simulations, especially those performed at constant pressure, that can
be difficult to catch.
For this reason, we set cellBasisVector3 to lz minus about 5 Å. Since we get about
55.9 Å for lz, line 13 of eq0.namd should read cellBasisVector3 0.0 0.0 51.0.
- 4
- While we have our system open in VMD, let's take a look at it.
Select Graphics
 Representations.... In the
Graphical Representations window, set Selected Atoms
to resname SIN to see only the
Representations.... In the
Graphical Representations window, set Selected Atoms
to resname SIN to see only the
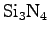 . Set the drawing method
Drawing Method to VDW. Now create a new
representation (by pressing Create Rep) with Selected
Atoms set to ions. The
. Set the drawing method
Drawing Method to VDW. Now create a new
representation (by pressing Create Rep) with Selected
Atoms set to ions. The
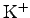 and
and
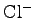 ions within the pore
should be visible. When you are finished examining the system, enter
mol delete all in the Tk Console.
ions within the pore
should be visible. When you are finished examining the system, enter
mol delete all in the Tk Console.
- 5
- To perform energy minimization, enter namd2 eq0.namd >
eq0.log in the terminal window. This may take a few minutes to
execute. During this time you may want to take a look at the next
step in the equilibration process eq1.namd. When the
minimization completes, check the end of log file eq0.log to
be certain that the simulation completed successfully.
| NAMD script |
steps |
description |
| eq0.namd |
201 |
energy minimization |
| eq1.namd |
500 |
raise temperature from 0 to 295 K, constant 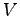 |
| eq2.namd |
1000 |
equilibrate, constant 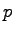 and Langevin thermostat and Langevin thermostat |
| run0.namd |
1000 |
apply 20 V, constant 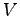 |
The table above summarizes the NAMD runs we will perform in this
section. It consists of three equilibration stages and one run with an
applied field. Stages such as these are used in most production
simulaions.
- 6
- Enter namd2 eq1.namd > eq1.log to gradually raise the
system's temperature from 0 K to 295 K at constant volume.
- 7
- Examine the NAMD configuration file eq2.namd in your
text editor. Notice the block of commands below the comment #
pressure control. These set the parameters for the Langevin piston
Nosé-Hoover method implemented in NAMD to maintain atmospheric
pressure. Close the text editor and equilibrate the system by entering
namd2 eq2.namd > eq2.log.
- 8
- Constant pressure simulations allow the volume of the system to
change. As a necessary condition for equilibrium, the volume should
fluctuate about a mean value. Select Extensions
 Analysis
Analysis  NAMD Plot from VMD's menu. In
the NAMD Plot window, select File
NAMD Plot from VMD's menu. In
the NAMD Plot window, select File  Select NAMD Log File, highlight eq2.log, and press Open. Select for VOLUME for the
Select NAMD Log File, highlight eq2.log, and press Open. Select for VOLUME for the 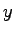 -axis data. Now, plot the system
volume versus time step by selecting File
-axis data. Now, plot the system
volume versus time step by selecting File  Plot
Selected Data. You should notice a significant downward trend in the
volume. At equilibrium, the volume fluctuates about a mean value for
an
Plot
Selected Data. You should notice a significant downward trend in the
volume. At equilibrium, the volume fluctuates about a mean value for
an 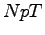 system such as this. Hence, we have not equilibrated long
enough. Since our time in this tutorial is limited, a system that has been equilibrated for 0.5 ns is included in this directory.
system such as this. Hence, we have not equilibrated long
enough. Since our time in this tutorial is limited, a system that has been equilibrated for 0.5 ns is included in this directory.
- 9
- We are now ready to apply an electric field and simulate the
flow of ionic current. Because the total current is more simply
related to voltage than the electric field magnitude, we are going to
apply a potential difference of 20 V along the
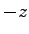 -axis of our
system. The corresponding uniform electric field is calculated by
-axis of our
system. The corresponding uniform electric field is calculated by
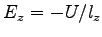 , where
, where 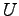 is the potential difference and
is the potential difference and 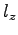 is the size of the system along the
is the size of the system along the 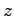 -axis. The NAMD unit for
electric field is
-axis. The NAMD unit for
electric field is
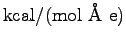 ; thus, the appropriate
conversion factor for
; thus, the appropriate
conversion factor for 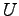 in V and
in V and 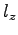 in Å is 23.0605492. That
is,
in Å is 23.0605492. That
is,
To obtain the value of 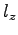 , open the NAMD extended system configuration file sample.xsc in your text editor.
Write down c_z, the tenth number in the row of system parameters.
Using
, open the NAMD extended system configuration file sample.xsc in your text editor.
Write down c_z, the tenth number in the row of system parameters.
Using
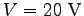 and
and
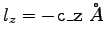 , calculate
, calculate
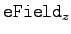 . Note that since the potential difference is applied the along
. Note that since the potential difference is applied the along 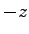 -axis,
-axis,
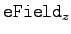 is positive.
is positive.
- 10
- Now open run0.namd. At the bottom of the file you will see the following lines:
| eFieldOn on |
|
| eField 0.0 0.0 0.0 |
|
Change the third component of eField to the value of
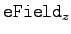 that you calculated. Before you close the
run0.namd, note that the pressure control lines are absent.
Applying an electric field to a pressure-controlled system will
distort it, leading to erroneous results. In addition, note that the
Langevin temperature control is only applied to the silicon nitride.
Applying Langevin forces to the ions, whose motion due to the electric
field we are trying to measure, could lead to a subtle bias in the
current.
that you calculated. Before you close the
run0.namd, note that the pressure control lines are absent.
Applying an electric field to a pressure-controlled system will
distort it, leading to erroneous results. In addition, note that the
Langevin temperature control is only applied to the silicon nitride.
Applying Langevin forces to the ions, whose motion due to the electric
field we are trying to measure, could lead to a subtle bias in the
current.
- 11
- Begin the simulation by entering namd2 run0.namd >
run0.log in the terminal window. The simulation may require a couple
minutes. Feel free to read ahead while it runs.
- 12
- We are using a very high applied electric field due to the time
constraints of this tutorial. If you analyze the temperature of the
simulation versus time step using the VMD plugin NAMD Plot
(whose use was described during the equilibration phase of this
section), you'll see that the temperature rises above 450 K, because
of the large ionic current. Such temperatures would render a
production simulation invalid. In real simulations, we would be using
a much smaller electric field.
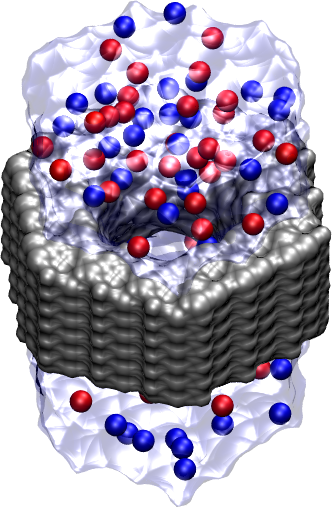
Figure 4:
Complete silicon nitride nanopore (grey) including water
and potassium (red) and chloride (blue) ions.
|
|
- 13
- Load the VMD save state by selecting File
 Load State... and then the file current.vmd. Step through
your trajectory and you should notice that the
Load State... and then the file current.vmd. Step through
your trajectory and you should notice that the
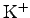 ions (in red)
move upward, while the
ions (in red)
move upward, while the
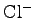 (in blue) ions move downward. Enter
mol delete all in the Tk Console.
(in blue) ions move downward. Enter
mol delete all in the Tk Console.
- 14
- The parameter dcdFreq is set to 100 in the NAMD
configuration file. As you may already know, this means that NAMD
writes the coordinates of every atom to a DCD file every 100
simulation steps. To calculate the ionic current, we will execute the
Tcl script electricCurrentZ.tcl. It computes the ionic
current by
where 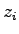 and
and 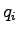 are respectively the
are respectively the 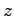 -coordinate and charge of ion
-coordinate and charge of ion 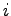 and
and
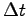 is the simulation time represented by dcdFreq.
Execute the script by entering source electricCurrentZ.tcl.
is the simulation time represented by dcdFreq.
Execute the script by entering source electricCurrentZ.tcl.
electricCurrentZ.tcl
# Calculate the current for a trajectory.
# Results are in "time(ns) current(nA)"
set dcdFreq 100
set selText "ions"
set startFrame 0
set timestep 1.0
# Input:
set pdb sample.pdb
set psf sample.psf
set dcd run0.dcd
set xsc run0.restart.xsc
# Output:
set outFile curr_20V.dat
# Get the time change between frames in femtoseconds.
set dt [expr $timestep*$dcdFreq]
# Read the system size from the xsc file.
# Note: This only works for lattice vectors along the axes!
set in [open $xsc r]
foreach line [split [read $in] "\n"] {
if {![string match "#*" $line]} {
set param [split $line]
puts $param
set lx [lindex $param 1]
set ly [lindex $param 5]
set lz [lindex $param 9]
break
}
}
puts "NOTE: The system size is $lx $ly $lz.\n"
close $in
# Load the system.
mol load psf $psf pdb $pdb
set sel [atomselect top $selText]
# Load the trajectory.
animate delete all
mol addfile $dcd waitfor all
set nFrames [molinfo top get numframes]
puts [format "Reading %i frames." $nFrames]
# Open the output file.
set out [open $outFile w]
#puts $out "sum of q*v for $psf with trajectory $dcd"
#puts $out "t(ns) I(A)"
for {set i 0} {$i < 1} {incr i} {
# Get the charge of each atom.
set q [$sel get charge]
# Get the position data for the first frame.
molinfo top set frame $startFrame
set z0 [$sel get z]
}
# Start at "startFrame" and move forward, computing
# current at each step.
set n 1
for {set f [expr $startFrame+1]} {$f < $nFrames && $n > 0} {incr f} {
molinfo top set frame $f
# Get the position data for the current frame.
set z1 [$sel get z]
# Find the displacements in the z-direction.
set dz {}
foreach r0 $z0 r1 $z1 {
# Compensate for jumps across the periodic cell.
set z [expr $r1-$r0]
if {[expr $z > 0.5*$lz]} {set z [expr $z-$lz]}
if {[expr $z <-0.5*$lz]} {set z [expr $z+$lz]}
lappend dz $z
}
# Compute the average charge*velocity between the two frames.
set qvsum [expr [vecdot $dz $q] / $dt]
# We first scale by the system size to obtain the z-current in e/fs.
set currentZ [expr $qvsum/$lz]
# Now we convert to nanoamperes.
set currentZ [expr $currentZ*1.60217733e5]
# Get the time in nanoseconds for this frame.
set t [expr ($f+0.5)*$dt*1.e-6]
# Write the current.
puts $out "$t $currentZ"
puts -nonewline [format "FRAME %i: " $f]
puts "$t $currentZ"
# Store the postion data for the next computation.
set z0 $z1
}
close $out
mol delete top
}
- 15
- The script electricCurrentZ.tcl produces an output file curr_20V.dat, which has two columns that record the time (ns) and the current (nA).
Open the file curr_20V.dat in a text editor.
Is the current steady? What is its mean value?



Next: Simulations of DNA permeation
Up: Bionanotechnology Tutorial
Previous: Introduction
Contents
www.ks.uiuc.edu/Training/Tutorials
![]() Representations....
In the Graphical Representations window, set the
Drawing Method to CPK. Now we can clearly see the
configuration of atoms in the unit cell. This configuration of eight
nitrogen atoms and six silicon atoms will form the basis for our
extended
Representations....
In the Graphical Representations window, set the
Drawing Method to CPK. Now we can clearly see the
configuration of atoms in the unit cell. This configuration of eight
nitrogen atoms and six silicon atoms will form the basis for our
extended
![]() crystal.
crystal.
![[*]](crossref.gif)
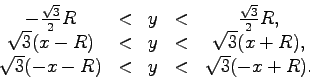
![\fbox{
\begin{minipage}{.2\textwidth}
\end{minipage} \begin{minipage}[r]{.75\te...
...\textit{Biophysical Journal} \textbf{87}, 2905--2911 (2004)).}
\end{minipage} }](img31.gif)
![\framebox[\textwidth]{
\begin{minipage}[r]{.75\textwidth}
\noindent\small\text...
...udegraphics[width=2.3 cm]{pictures/ypore.png}}
\end{minipage}}
\end{minipage} }](img32.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\end{minipage} \begin{min...
...es
its absolute value, which is negligible for most purposes.}
\end{minipage} }](img33.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\end{minipage} \begin{min...
...e {\tt chargeSi} in the PSF
generating script (See Appendix).}
\end{minipage} }](img37.gif)
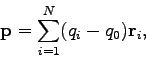
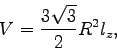
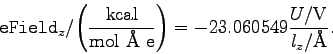
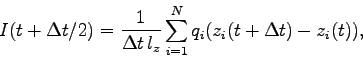
![\framebox[\textwidth]{
\begin{minipage}[r]{.75\textwidth}
\noindent\small\text...
...s section. How does
the current compare to double-cone pore? }
\end{minipage} }](img70.gif)
![\fbox{
\begin{minipage}{.2\textwidth}
\end{minipage} \begin{minipage}[r]{.75\te...
... a nanopore can be used to detect
and study single molecules.}
\end{minipage} }](img71.gif)