


Next: Getting Started
Up: AARS Tutorial
Previous: Contents
Contents
Subsections
The new Multiple Sequence Alignment version of VMD that is currently under development was originally
created to allow biomedical researchers to study the evolutionary changes in sequence and
structure of proteins across all three domains of life, from bacteria to humans. The comparative sequence and structure metrics, and
analysis tools introduced in the accompanying article by O'Donoghue and Luthey-Schulten
1
are now part of this new version. In particular, the Luthey-Schulten group has included
a recently developed structure-based measure of homology
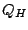 (see Appendix B), that takes into account the
effect of insertions and deletions and has been shown to produce accurate structure-based
phylogenetic trees. The STAMP structural alignment algorithm, kindly provided by our colleagues Russell and Barton, is included in our
alpha release 2 . We plan to offer biomedical researchers a tool to examine the changes in protein structure
in the correct statistical framework. As a result, Multiple Sequence Alignment is an invaluable tool for relating protein structure to its function or misfunction.
Since the accompanying tutorials were created for a program that is truly a Òwork in
progress,Ó we limit our demonstrations to the examination only of the correlation of
sequence and structure changes and represent these changes in terms of structural phylogenetic
trees.
(see Appendix B), that takes into account the
effect of insertions and deletions and has been shown to produce accurate structure-based
phylogenetic trees. The STAMP structural alignment algorithm, kindly provided by our colleagues Russell and Barton, is included in our
alpha release 2 . We plan to offer biomedical researchers a tool to examine the changes in protein structure
in the correct statistical framework. As a result, Multiple Sequence Alignment is an invaluable tool for relating protein structure to its function or misfunction.
Since the accompanying tutorials were created for a program that is truly a Òwork in
progress,Ó we limit our demonstrations to the examination only of the correlation of
sequence and structure changes and represent these changes in terms of structural phylogenetic
trees.
This tutorial showcases the new software tools in Multiple Sequence Alignment and will allow the reader to reconstruct
the figures in the accompanying review article entitled ``The Evolution of Structure in Aminoacyl-tRNA Synthetases.''
It is designed such that it can be used by both new and previous users of VMD, however,
it is highly recommended that new users go through the ``VMD Molecular Graphics'' tutorial
in order to gain a working knowledge of the program. This tutorial has been designed
specifically for VMD with Multiple Sequence Alignment and should take about an hour to complete in its entirety.
The aminoacyl-tRNA synthetases (AARSs) are key proteins involved in
the translation machinery in living organisms; it is not surprising, therefore, that these enzymes are found
in all three domains of life. There are twenty specific tRNA synthetases (one for each amino acid), although not all
organisms contain the full set. Studying the function, structure, and evolution of these proteins remains an area of intense interest as, in addition
to being a major constituent of the translation process, these proteins are also believed to contain vital information spanning the evolution of life
from the ancient ``RNA world'' to the modern form of life.
Figure 1:
The reaction catalyzed by the aminoacyl tRNA synthetases (aa could be any amino acid).
|
|
The AARSs are responsible for loading the twenty different amino acids onto the cognate tRNA during protein synthesis (see Figure 1).
Each AARS is a multidomain protein consisting of (at least) a catalytic domain and an anticodon binding domain. In all known cases,
the synthetases divide into class I or class II types; class I AARSs exemplify the basic Rossmann fold, while class II AARSs exhibit a
fold that is unique to them and biotin synthetase holoenzyme.
Additionally, some of the AARSs, for example aspartyl-tRNA synthetase, have an ``insert domain'' within their
catalytic domain (see Figure 2). Recognition of the tRNA molecule is typically performed by the anticodon domain, however
residues that have degenerate codons (e.g. serine has six different codons) have been found to exploit other
features in the tRNA for recognition (e.g. the acceptor arm or the so-called discriminator base).
These molecular machines operate with remarkable precision, making only one mistake in every 10,000
translations. The intricate architecture of specific tRNA synthetases helps to discriminate against mis-coding.
Figure 2:
A snapshot of AspRS-tRNA aspartyl-adenylate complex (from E. Coli) in the active form. Note the anticodon binding domain (orange),
the insertion domain (pink), and the catalytic domain (blue). tRNA is docked to AspRS, and the catalytic active site is highlighted
within the catalytic domain (red bubble); the aspartyl-adenylate substrate is shown in space-filling representation. The residues involved in specific base
recognition on the tRNA are also highlighted within the anticodon binding domain (green bubble). Note that specific contacts between the
tRNA and Asp-RS allow for strategic positioning of the tRNA relative to the enzyme.
|
|



Next: Getting Started
Up: AARS Tutorial
Previous: Contents
Contents
workshop@ks.uiuc.edu

