

Next: Scripting in VMD
Up: VMD Tutorial
Previous: Working with a Single
Subsections
Trajectories and Movie Making
The time-evolving coordinates of a system are called a trajectory.
They are most commonly obtained in simulations of molecular systems,
but can also be generated by other means and for different purposes.
Upon loading a trajectory, VMD can animate movies of
system evolving with time and analyze various structural
features throughout the trajectory.
This section will introduce the basics of working with trajectory
data in VMD.
You will also learn how to analyze trajectory data in
Section 4.
Loading Trajectories
Trajectory files are typically large binary files that contain
the time varying atomic coordinates for the system.
Each set of coordinates corresponds to one frame in time.
An example of a trajectory file is a DCD file.
Trajectory files usually do not contain information structural information
as found in protein structure files (PSF). Therefore, we must first
load the structure file, and then add the trajectory data to the
same molecule, so that VMD has access to both the structure and trajectory
information.
- 1
- Start a new VMD session. In the VMD Main window, select File
 New Molecule.... The Molecule File Browser window should appear on your screen.
New Molecule.... The Molecule File Browser window should appear on your screen.
- 2
- Use the Browse... button to find the file ubiquitin.psf in vmd-tutorial-files in the tutorial directory. When you select the file, you will be back in the Molecule File Browser window. Press the Load button to load the molecule.
- 3
- In the Molecule File Browser window, make sure that ubiquitin.psf is selected in the Load files for: pull-down menu on the top, and click on the Browse button. Browse for pulling.dcd. Note the options available in the Molecule File Browser window: one can load trajectories starting and finishing at chosen frames, and adjust the stride between the loaded frames. Leave the default settings so that the whole trajectory is loaded.
- 4
- Click on the Load button in the Molecule File Browser window. You will be able to see the frames as they are loaded into the molecule. After the trajectory finishes loading, you will be looking at the last frame of your trajectory. To go to the beginning, use the animation tools at the lower part of the VMD Main menu (see Fig. 17). You can close the Molecule File Browser window.
Figure 17:
Animation tools in the main menu of VMD. The tools allow one to go over frames of the trajectory (e.g., using the
slider) and to play a movie of the trajectory in various modes (Once, Loop, or Rock) and at an adjustable speed.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.0\textwidth]{FIGS/vmd-animation-tools}
}
\end{center}
\end{figure}](img39.gif) |
- 5
- For a convenient visualization of the protein, choose Graphics
 Representations in the VMD Main menu. In the Selected Atoms field, type protein and hit Enter on your keyboard; in the Drawing Method, select NewCartoon; in the Coloring Method, select Structure.
Representations in the VMD Main menu. In the Selected Atoms field, type protein and hit Enter on your keyboard; in the Drawing Method, select NewCartoon; in the Coloring Method, select Structure.
The trajectory you just loaded is a simulation of an AFM (Atomic Force Microscopy) experiment pulling on a single ubiquitin molecule, performed using the Steered Molecular Dynamics (SMD) method (Isralewitz et al., Curr. Opin. Struct. Biol., 11:224, 2001). We are looking at the behavior of the protein as it unfolds while being pulled from one end, with the other end being constrained to its original position. Each frame step corresponds to 10ps. Ubiquitin has many functions in the cell, and it is currently believed that some of these functions depend on the protein's elastic properties. Such elastic properties are usually due to hydrogen bonding between residues in 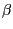 strands of the protein molecules.
strands of the protein molecules.
Main Menu Animation Tools
You can now play the movie of the loaded trajectory back and forth, using the animation tools in Fig. 17.
- 1
- By dragging the slider one navigates through the trajectory. The buttons to the left and to the right from the slider panel allow one to jump to the end of the trajectory or go back to the beginning.
- 2
- For example, create another representation for water in the Graphical Representations window: click on the Create Rep button; in the Selected Atoms field, type water and hit enter; in Drawing Method, choose Lines; in the Coloring Method, select Name. This shows the water droplet present in the simulation to mimic the natural environment for the protein. Using the slider, observe the behavior of the water around the protein. The shape of the water droplet changes throughout the simulation, because water molecules follow the protein as it unfolds, due to interactions with the protein surface.
- 3
- When playing animations, you can choose between three looping styles: Once, Loop and Rock. You can also jump to a frame in the trajectory by entering the frame number in the window on the left of the slider panel.
Trajectory Visualization
We will now learn some basic visualization tricks that are useful for working with trajectories.
- 1
- For clarity, turn off the water representation by double-clicking on it in the Graphical Representations window. As you might have noticed, when we play the animation, the protein movements are not very smooth due to thermal fluctuations (as the simulation is performed under the conditions that mimic a thermal bath).
- 2
- VMD can smooth the animation by averaging some number of frames. In the Graphical Representations window, select your protein representation and click the Trajectory tab. At the bottom, you see Trajectory Smoothing Window Size set to zero. As your animation is playing, increase this setting. Notice that
the motion gets smoother and smoother as the setting goes up.
We will learn now how to display many frames of the same trajectory at once.
- 3
- In the Graphical Representations window, highlight your protein representation by clicking on it and press the Create Rep button. This creates an identical representation, but note that smoothing is set to zero. Hide the old protein representation.
- 4
- Highlight the new protein representation and click the Trajectory tab. Above the smoothing control, notice the Draw Multiple Frames control. It is set to now by default, which is simply the current frame. Enter 0:10:99, which selects every tenth frame from the range 0 to 99.
- 5
- Go back to the Draw style tab, and select Coloring Method
 Trajectory
Trajectory
 Timestep.
This will draw the beginning of the trajectory in red, the middle in white, and the end in blue.
Timestep.
This will draw the beginning of the trajectory in red, the middle in white, and the end in blue.
- 6
- We can also use smoothing to make the large-scale motion of the protein more apparent. Go back to the Trajectory tab, set the smoothing window to 20. The result should be similar to Fig. 18.
Figure 18:
Image of every tenth frame showed at once, smoothed with a 20-frame window.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=0.50\textwidth]{FIGS/allframes}
}
\end{center}
\end{figure}](img40.gif) |
Now we will see how to make VMD update the selection each frame.
- 7
- Hide the current representation showing all frames, and display only the water representation by double-clicking on it. Change the text in the Selected Atoms from water to water and within 3 of protein and hit enter. This will show all water atoms within 3Å of the protein.
- 8
- Play the trajectory. Although the displayed water atoms may be near the protein for a little while, they soon wander off, and are still shown despite no longer meeting the selection criteria. The Update Selection Every Frame option in the Trajectory tab of the Graphical Representations window remedies this. If the option box is checked, the selection is updated every frame. See Fig. 19.
- 9
- Quit VMD.
Figure 19:
Water within 3 Å of the protein, shown for a selection that is not updated and for the one that is updated each
frame. The snapshots shown are (from left to right) for frames 0, 17, and 99.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.0\textwidth]{FIGS/update-selection}
}
\end{center}
\end{figure}](img41.gif) |
The Basics of Move Making in VMD
We will now learn how to make a basic movie.
- 1
- Start a new VMD session. Repeat steps 1-5 in Section 2.1 to load the ubiquitin trajectory into VMD and display the protein in a secondary structure representation.
- 2
- To make movies, we will use the VMD Movie Maker plugin. In the VMD Main window, go to menu item Extensions
 Visualization
Visualization
 Movie Maker. The VMD Movie Generator window will appear (Fig. 20).
Movie Maker. The VMD Movie Generator window will appear (Fig. 20).
- 3
- First, let us look at some of the options for making a movie. Click on the Movie Settings menu in the VMD Movie Generator window. You can see that in addition to a trajectory movie, Movie Maker can also make a movie by rotating the view point of a single frame. In the Renderer menu, one can choose the type of renderer for making the movie. We will use the default option, Snapshot. One can also choose the output file format for the movie in menu item Format.
- 4
- We will first make a movie of just one frame of the trajectory. For that purpose, select Rock and Roll option in the Movie Settings menu in the VMD Movie Generator window. Set the working directory to any convenient directory of your choice, give your movie a name, and click Make Movie.
- 5
- Once rendering is finished, open and view the movie with your favorite application. This movie setting is good for showing one side of your system primarily.
- 6
- Now we will make a movie of the trajectory. In the VMD Movie Generator window, select Movie Settings
 Trajectory, give this one a different name, and click Make Movie. Note that the length of the movie is automatically set 24 frames per second. For a trajectory, duration of the movie can be decreased, but cannot be increased.
Trajectory, give this one a different name, and click Make Movie. Note that the length of the movie is automatically set 24 frames per second. For a trajectory, duration of the movie can be decreased, but cannot be increased.
Figure 20:
The VMD Movie Generator window.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=0.6\textwidth]{FIGS/movie-maker}
}
\end{center}
\end{figure}](img44.gif) |
- 7
- Try out different options in the VMD Movie Generator window. Once you are done, quit VMD.


Next: Scripting in VMD
Up: VMD Tutorial
Previous: Working with a Single
vmd@ks.uiuc.edu
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.0\textwidth]{FIGS/vmd-animation-tools}
}
\end{center}
\end{figure}](img39.gif)
![]() strands of the protein molecules.
strands of the protein molecules.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[width=1.0\textwidth]{FIGS/update-selection}
}
\end{center}
\end{figure}](img41.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
...vie file is determined by the size (resolution) of that window.}
\end{minipage}}](img42.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2.5...
... page at http://www.ks.uiuc.edu/Research/vmd/plugins/vmdmovie/.}
\end{minipage}}](img43.gif)