

Up: VMD Images and Movies
Previous: Working with Still Frames
Subsections
Working With Trajectories
In the second unit, you will learn how to work with
trajectories.
In this section, we will learn how to smooth trajectories, show
multiple frames at once, and make atom selections ``follow'' the
trajectory.
- 1
- Open VMD, and load the files dna.psf and trajectory.dcd. Before clicking Load in the
Molecular File Browser window, look at the Frames section.
Here you have precise control over how the file is loaded. Leave them
at the default setting, so that all frames of the trajectory are
loaded.
- 2
- Set up a representation showing only the DNA, in the NewCartoon drawing style.
- 3
- Play the animation, as you learned how to do in the VMD
Tutorial. You may want to lower the speed!
- 4
- The movement is not very smooth, due to thermal
fluctuations. VMD can smooth the animation by averaging some number of
frames. In the Graphical Representations window, select your
DNA representation, and click the Trajectory tab. At the
bottom, you see Trajectory Smoothing Window Size set to
zero. As your animation is playing, increase this setting. Notice that
the motion gets smoother and smoother as you go up.
- 5
- Now we'll see how to display many frames at once. Create a new
DNA representation, and hide the old one. (Hint: Select the old
representation before hitting the Create Rep button, so that
the new one already has the right selection and drawing style. Note that
smoothing will be set to zero.)
- 6
- Click the Trajectory tab again. Above the smoothing
control, notice the Draw Multiple Frames control. It is set
to now by default, which is simply the current frame. Enter 0:10:199, which selects every tenth frame from the range 0 to 199.
- 7
- Go back to the Draw style tab, and change the
coloring mode to Timestep. This will draw the beginning of
the trajectory in red, the middle in white, and the end in blue. Also
set Material to Transparent.
- 8
- We can also use smoothing, making the large-scale motion of the
DNA more apparent. Set the smoothing window to 10. You should have
something like Fig. 10(a).
Figure 10:
Two examples of showing multiple frames at once.
[Image of every tenth frame showing at once, smoothed with a ten-frame window.] ![\includegraphics[scale=0.75]{pictures/u2-allframes-2}](img27.gif)
[Image of all frames showing at once, for two Na  ions and two Cl  ions. Water is shown as a transparent MSMS surface.] ![\includegraphics[scale=0.75]{pictures/u2-allframes-ions-2}](img28.gif)
|
- 9
- Finally, we'll see how to make VMD ``update'' the selection
each frame. Hide the representation showing all frames, and reshow
your first representation, showing only DNA. Then make a new
representation, with the selection text
ions and within 5 of segname ADNA
This will show all ions within 5 Å of the DNA.
Figure 11:
Movie noupdate.mpg.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.85]{pictures/u2-noupdate-2-frames}
}
\end{center}
\end{figure}](img29.gif) |
- 10
- Now play the trajectory. (A movie of the trajectory is provided
as noupdate.mpg, and illustrated in Fig. 11.)
Notice anything wrong? Although the ions may be near the DNA for a
little while, they soon wander off, and are still shown despite no
longer meeting the selection criteria. The Update Selection
Every Frame option in the Trajectory tab of the
Graphical Representations window remedies this, doing exactly what
its name says. See Fig. 12
Figure 12:
Movie update.mpg.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.85]{pictures/u2-update-frames}
}
\end{center}
\end{figure}](img30.gif) |
You now know the basics of working with trajectories. We will
next look at some more advanced things we can do with them.
In this section, we will explore an advanced technique for
visualization. We will look at a trajectory from a simulation in which
a ``virtual pore'' was collapsed around a short strand of DNA, and see
how to use VMD to superimpose the position of the virtual pore at each
timestep. Along the way we will learn about VMD's custom graphics
capabilities.
- 1
- Start a fresh VMD session, and load the files pore.psf and pore.dcd, and set up a representation showing
just the DNA.
- 2
- For this section, we will be examining a script. Let's see what
it does. Type the following in the Tk Console window:
| source pore.tcl |
|
| enabletrace |
|
- 3
- Start the animation. The blue cylinder is drawn at the radius
of the potential used in the simulation. (See Fig. 13.)
Figure 13:
Movie pore.mpg.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.85]{pictures/u2-pore-2-frames}
}
\end{center}
\end{figure}](img32.gif) |
- 4
- Now hide your DNA representation, and type the command enablearrows.
- 5
- Restart the animation. The DNA is now drawn by the script as
well, represented as arrows. You should see something like
Fig. 14. The command disablearrows will turn this
off. disabletrace will turn off everything done by this
script.
Figure 14:
DNA represented as arrows.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=1.0]{pictures/u2-arrows-2}
}
\end{center}
\end{figure}](img33.gif) |
- 6
- Now that we see what it does, let's look at the script. Open
the file pore.tcl in a text editor.
- 7
- The first few lines defines the procs for enabling and disabling
the script:
| proc enabletrace {} { |
|
| global vmd_frame |
|
| trace variable vmd_frame([molinfo top]) w drawcounter |
|
| } |
|
| proc disabletrace {} { |
|
| global vmd_frame |
|
| trace vdelete vmd_frame([molinfo top]) w drawcounter |
|
| } |
|
These procs use the trace feature of Tcl, which is a mechanism
for setting up some action when some variable changes. The command
trace variable vmd_frame([molinfo top]) w drawcounter
says to call the command drawcounter everytime the variable
vmd_frame
([molinfo top]) is written (w above is short
for write). See the VMD User's Guide at
http://www.ks.uiuc.edu/Research/vmd/current/ug/ug.html
for a list of
VMD variables that trace can be used with.
- 8
- The next few lines set up the procs to turn the arrows on and off:
| proc enablearrows {} { |
|
| global pore_draw_arrows |
|
| set pore_draw_arrows 1 |
|
| } |
|
| proc disablearrows {} { |
|
| global pore_draw_arrows |
|
| set pore_draw_arrows 0 |
|
| } |
|
- 9
- Next we have procs which draw the arrows:
| proc vmd_draw_arrow_BASE {mol start end} { |
|
| set coneEnd [vecadd $end [vecscale 0.25 [vecsub $end $start]]] |
|
| graphics $mol cylinder $start $end radius 1.0 |
|
| graphics $mol cone $end $coneEnd radius 1.5 |
|
| } |
|
| proc vmd_draw_arrow {mol start end} { |
|
| set middle [vecadd $start [vecscale 0.8 [vecsub $end $start]]] |
|
| graphics $mol cylinder $start $middle radius 1.0 |
|
| graphics $mol cone $middle $end radius 1.5 |
|
| } |
|
These simply draw the arrows by using built-in procs for drawing
cylinders and cones.
- 10
- Next, the script sets some parameters defining the size of the
pore cylinder and how it changes through the trajectory:
| set Rstart 14 |
|
| set Rrate 0.00001 |
|
| set Rtarget 4.0 |
|
| set frameRate 10000 |
|
| set Center 0.0 |
|
Obviously, these should coincide with those used in the original
simulation.
- 11
- Now we make note of the DNA residues:
| set sel [atomselect top "segid ADNA and name P"] |
|
| set residList [lsort -integer [$sel get resid] ] |
|
| set residStart [lindex $residList 0] |
|
| set residEnd [lindex $residList end] |
|
Figure 15:
Movie arrows.mpg.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.85]{pictures/u2-arrows-2-frames}
}
\end{center}
\end{figure}](img34.gif) |
- 12
- Now we get to the real meat of the script: the drawcounter proc. This is the procedure called every time the frame
changes.
| proc drawcounter { name element op } { |
|
| ... |
|
| } |
|
trace calls the callback proc with three aruments, which are
basically just the arguments we provided trace to begin with,
and which we don't use.
- 13
- Now let's look at the drawcounter proc itself. First, it
sets some global variables, which we won't reproduce here. Then it
calculates the proper radius of the cylinder for the current timestep:
set R [expr $Rstart - $Rrate*$frameRate 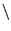 |
|
| *$vmd_frame([molinfo top]) + 1.0] |
|
| if {$R < $Rtarget} { |
|
| set R $Rtarget |
|
| } |
|
- 14
- Next is the code which actually draws the cylinder:
| draw delete all |
|
| draw color blue |
|
| draw material Transparent |
|
| set start_pt [list 0 0 [expr $Center - 30]] |
|
| set stop_pt [list 0 0 [expr $Center + 50]] |
|
| draw cylinder $start_pt $stop_pt radius $R resolution 32 |
|
Here we use VMD's draw command to set the drawing parameters
and draw the cylinder.
- 15
- Finally, we draw the DNA arrows, if it's enabled:
| if { $pore_draw_arrows } { |
|
| for {set resid [expr $residStart]} {$resid < $residEnd} {incr resid} { |
|
| set residONE [expr $resid + 1] |
|
| set tmpSel1 [atomselect top "resid $resid and name N1 C3'" |
|
| frame $vmd_frame([molinfo top])] |
|
| foreach {coordN1 coordC1} [$tmpSel1 get "x y z"] { break } |
|
| draw color yellow |
|
| vmd_draw_arrow_BASE top $coordC1 $coordN1 |
|
| set tmpSel2 [atomselect top "(resid $resid and name P) |
|
| or (resid $residONE and name P)" frame $vmd_frame([molinfo top])] |
|
| foreach {coordP1 coordP2} [$tmpSel2 get "x y z"] { break } |
|
| draw color 14 |
|
| vmd_draw_arrow top $coordP1 $coordP2 |
|
| $tmpSel1 delete |
|
| $tmpSel2 delete |
|
| } |
|
| } |
|
The script simply uses the atomic coordinates of three of the atoms of
each base to define where the arrow should be drawn, then the
appropriate drawing command, defined above, is called.
You now have some powerful techniques for working with
trajectories. The last section of this tutorial will show you how to
take everything you've learned to produce movies of your system.
In this final section, you'll first learn how to make a basic
movie. Then we'll see how to do more advanced animations, again using
Tcl's callback functionality.
- 1
- Leave everything loaded in VMD from the last section, or start
a fresh VMD session and type source makingmovies-restart.tcl in
the Tk Console window to load everything you need for this
section. To make our movies, we will use the VMD Movie Maker
plugin. Go to Extensions
 Visualization
Visualization
 Movie Maker.
Movie Maker.
- 2
- First, let's look at some of the options for our movie. Click
on the Movie Settings menu. In additon to a trajectory movie,
Movie Maker can also take still frames and animate camera motion.
- 3
- Set up your representations and angle to something you like.
- 4
- Select Rock and Roll, set the working directory to
something convenient, give your movie a name, and click Make
Movie. You will see the VMD display update as the movie is made.
- 5
- Once VMD is finished, open the movie. This movie setting is
good for showing one side of your system primarily.
- 6
- Now, try a trajectory movie. Select Movie Settings
 Trajectory, give this one a different name, and click
Make Movie.
Trajectory, give this one a different name, and click
Make Movie.
- 7
- It is possible to show multiple views of your molecule at once,
which can be very helpful in some cases. Load the files pore.psf and pore.dcd again, creating a new molecule. (You
may want to rename it by double-clicking its name.)
- 8
- Fix the original molecule by double-clicking the ``F'' next
to its name in the VMD Main window. Now you can manipulate
the view of the second copy without affecting the first. Make your
window wider, and use the Translate mouse mode to shift your
second view to the left of the first.
Figure 16:
Movie 2views.mpg.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.85]{pictures/u2-2views-2-frames}
}
\end{center}
\end{figure}](img37.gif) |
- 9
- Since the new copy is its own molecule, you have complete
control of how it is displayed, independent of the original copy. When
you play the trajectory or make a movie, VMD will keep the two views
synchronized. The file 2views.mpg shows an example of this
technique, as does Fig. 17.
Figure 17:
Example of showing the same object from multiple
perspectives.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.75]{pictures/u2-2views}
}
\end{center}
\end{figure}](img38.gif) |
- 10
- Finally, we'll look at how to use user-defined scripts to
dictate the animation. Type the following commands in the Tk
Console:
| source usermovie.tcl |
|
| enablemoviecallback |
|
Now select Movie Settings  User Defined
Procedure, and click Make Movie. Now, in addition to simply
going through the trajectory, the camera starts above the DNA and
rotates down and around it. An example movie is provided, and shown in
Fig. 18
User Defined
Procedure, and click Make Movie. Now, in addition to simply
going through the trajectory, the camera starts above the DNA and
rotates down and around it. An example movie is provided, and shown in
Fig. 18
Figure 18:
Movie usermovie.mpg
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.85]{pictures/u2-usermovie-frames}
}
\end{center}
\end{figure}](img39.gif) |
- 11
- Let's see how it works. Its contents are reproduced here:
| set trajectory_frames [molinfo top get numframes] |
|
| proc moviecallback { args } { |
|
| global trajectory_frames |
|
| set frac [expr double($::MovieMaker::userframe)/$::MovieMaker::numframes] |
|
| rotate z to [expr $frac*360.0] |
|
| rotate x by [expr -90.0*$frac] |
|
| set frac [expr double($::MovieMaker::userframe)/$::MovieMaker::numframes] |
|
| animate goto [expr $frac * $trajectory_frames] |
|
| } |
|
| proc enablemoviecallback { } { |
|
| animate goto 0 |
|
| trace add variable ::MovieMaker::userframe write moviecallback |
|
| } |
|
| proc disablemoviecallback { } { |
|
| trace remove variable ::MovieMaker::userframe write moviecallback |
|
| } |
|
This should look familiar. We are again using the trace feature
of Tcl. The Movie Maker plugin sets the writes the variable ::MovieMaker::userframe each time it changes frames for animation,
and when that happens, our proc moviecallback is called. This
proc rotates the scene, and advances the animation frame.
You've now completed the tutorial. We've seen a number of
techniques for representing molecular systems clearly and effectively,
and gotten a taste of VMD's capabilities. The VMD website, listed at
the beginning of this tutorial, has many other tutorials and resources
for VMD users.


Up: VMD Images and Movies
Previous: Working with Still Frames
tutorial-l@ks.uiuc.edu
![\includegraphics[scale=0.75]{pictures/u2-allframes-2}](img27.gif) [Image of all frames showing at once, for two Na
[Image of all frames showing at once, for two Na![\includegraphics[scale=0.75]{pictures/u2-allframes-ions-2}](img28.gif)
![\includegraphics[scale=0.75]{pictures/u2-allframes-2}](img27.gif) [Image of all frames showing at once, for two Na
[Image of all frames showing at once, for two Na![\includegraphics[scale=0.75]{pictures/u2-allframes-ions-2}](img28.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...ultiple trajectory files into one, with
a variety of options.}
\end{minipage} }](img31.gif)
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...y the different
rendering options in the {\sf Renderer} menu.}
\end{minipage} }](img36.gif)
![]() User Defined
Procedure, and click Make Movie. Now, in addition to simply
going through the trajectory, the camera starts above the DNA and
rotates down and around it. An example movie is provided, and shown in
Fig. 18
User Defined
Procedure, and click Make Movie. Now, in addition to simply
going through the trajectory, the camera starts above the DNA and
rotates down and around it. An example movie is provided, and shown in
Fig. 18