



Next: Summary of
Results
Up: Residue Selection
Previous: Exercise 2:
Sequence Conservation Contents
This exercise will further examine structure conservation. Make sure to
change Molecule Coloring
back to Q per residue.
- 1 Go back to the main MultiSeq window top pull-down
menu and select Tools
 Residue Selection. Residue Selection.
- 2 Select Q Value.
- 3 Select Less than and 0.1 value.
- 4 Then click on the Select button.
|

|
If you had set Molecule Coloring to Q per residue you
will notice that the majority of the red areas have turned yellow, and
the blue is visible. If you look in the Sequence Display, you
will see that the majority of SerRS residues have been highlighted. By
using these settings in Q value, you have demonstrated where the
aligned molecules have the least structural conservation,Q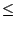 0.1.
0.1.
Figure 19:
Q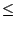 0.1
0.1

|
Now select Greater than or equal to and 0.6 in the Q
Value window , and click on Select. Notice, in the OpenGL
Display that the majority of the Blue area inside the molecules has
turned yellow. Also, note what has occurred in the sequence display.
Many areas of the AspRS molecules are highlighted. Occasionally, an
amino acid in SerRS is highlighted along with those of the AspRS. As
you increase the value to 0.7, there are no highlighted residues from
SerRS. Now that we have completed the exercises in Residue Selection,
close the
Select window.




Next: Summary of
Results
Up: Residue Selection
Previous: Exercise 2:
Sequence Conservation Contents
Brijeet Dhaliwal
2004-09-15









