
Next: Output: Water Sphere Log
Up: Basics of NAMD
Previous: Ubiquitin in a Water
Subsections
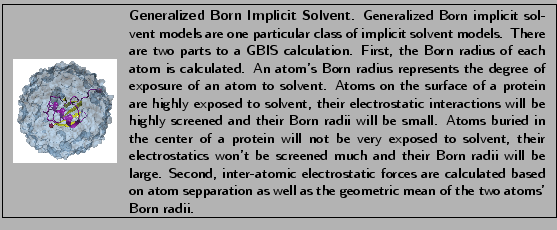
Ubiquitin in Generalized Born Implicit Solvent
In this section, you will examine the minimization and equilibration of ubiquitin in generalized Born implicit solvent (GBIS).
- 1
- Go to your 1-4-gbis directory by typing cd ../1-4-gbis . Here, you will find a configuration file for the minimization and equilibration of ubiquitin in implicit solvent. All output files for simulation of your ubiquitin in implicit solvent will be placed in this directory.
- 2
- Open the configuration file, ubq_gbis_eq.conf by typing VMD text editor ubq_gbis_eq.conf.
The configuration file contains some commands which are different than the water sphere and periodic boundary conditions configuration files. Here, the differences are pointed out and explained.
- 3
- Because there is currently no way of calculating long-range interactions in implicit solvent, such as through PME, features based on periodic boundary conditions are not used with GBIS as listed below.
- Periodic Boundary Conditions are not used.
- PME is not used.
- Constant Pressure Control (variable volume) is not used.
- 4
- A few additional commands are required for implicit solvent use. The new commands are listed:
- structure & coordinates: implicit solvent simulations use structures which do not include any explicit water or ions as these are represented implicitly.
- gbis: indicates whether or not the simulation uses the generalized Born implicit solvent model. Default value is no; set to yes to utilize.
- cutoff: because there is no long-range electrostatic calculation, cutoff should be set higher for GBIS than for PME simulations; a value of 14 Å is sufficient here.
- alphaCutoff: sets the cutoff used to determine the Born radius of each atom. It is reasonable to set it a few Ås less than cutoff; alphaCutoff = 12 Å should be sufficient here.
- ionConcentration: sets the concentration of implicit ions. Increasing the ion concentration increases the electrostatic screening; the default value is 0.2 M.

Your implicit solvent simulations should take roughly the same time as the water sphere simulation. The simulation will produce output files similar to the ubiquitin in water sphere simulation, which are discussed in Section 1.7.


Next: Output: Water Sphere Log
Up: Basics of NAMD
Previous: Ubiquitin in a Water
namd@ks.uiuc.edu