

Next: Other bioinformatics tools
Up: Bioinformatics Tutorial
Previous: Viewing conserved domains of
Molecular phylogenetic tree.
In this section you will plot a dendrogram displaying the measured similarities between the seven proteins which you pairwise aligned in Section 5. You will compare there relative position in the dendrogram to their relative position in the phylogenetic tree of life.
Below is the pairwise alignment scores from the 21 pairs aligned in
section 5. The information in ~/tbss.work/Bioinformatics/multipleData/stats is
assembled into a symmetric matrix:
| |
1EQR |
1ATI |
1ADJ |
1EFW |
1ASZ |
1B8A |
1BBW |
| 1EQR |
0.0 |
0.21 |
0.23 |
0.55 |
0.28 |
0.31 |
0.31 |
| 1ATI |
0.21 |
0.0 |
0.25 |
0.24 |
0.24 |
0.18 |
0.21 |
| 1ADJ |
0.23 |
0.25 |
0.0 |
0.24 |
0.24 |
0.21 |
0.23 |
| 1EFW |
0.55 |
0.24 |
0.24 |
0.0 |
0.34 |
0.36 |
0.30 |
| 1ASZ |
0.28 |
0.24 |
0.24 |
0.34 |
0.0 |
0.41 |
0.27 |
| 1B8A |
0.31 |
0.18 |
0.21 |
0.36 |
0.41 |
0.0 |
0.28 |
| 1BBW |
0.31 |
0.21 |
0.23 |
0.30 |
0.27 |
0.28 |
0.0 |
The commands in the following Matlab session are all in the Matlab script Dendro.m. The commands can be run all at once simply by typing Dendro
at the Matlab command line when Matlab's current directory contains Dendro.m.
Now we will use the clustering algorithms in the Statistics toolbox of Matlab
to draw a dendrogram of the relatedness of the domains. Here we use the above
scores derived from sequence alignment, but structure alignment scores could
be used as well[4].
- Move to the directory for this exercise with cd ~/tbss.work/Bioinformatics/matlabData
- Start Matlab by typing at the UNIX console: matlab.
- The commands in the following Matlab session are all in the Matlab script
Dendro.m. The commands can be run all at once simply by typing Dendro at the Matlab command line, as long as Matlab's current directory contains Dendro.m and distM.dat. If you like, type in the below, or paste lines into
the Matlab command line from Dendro.m or the web-based version of this
tutorial. (To see the numerical result of a calculation, leave of the semicolon
from the end of the line. To see the value of a variable, enter its name
alone on the Matlab command line.)
- First, we read in the above distance matrix of sequence similarity for 7 proteins.
load distM.dat;
- We make a new matrix by subtracting the sequence similarity values from 1,
so that longer distances in our dendrogram will correspond to greater
evolutionary distance.
dM=1-distM;
- Its important to keep track of names of the proteins...
l={'1eqr','1ati','1adj','1efw','1asz','1b8a','1bbw'};
- To use the 'linkage' command of Matlab, one needs to form a column vector of
the
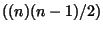 non-redundant elements above the main diagonal of the
non-redundant elements above the main diagonal of the
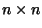 distance matrix; our
distance matrix; our 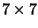 matrix produces a 21-element vector:
matrix produces a 21-element vector:
d=[dM(2:7,1);dM(3:7,2);dM(4:7,3);dM(5:7,4);dM(6:7,5);dM(7:7,6)];
- Use the linkage command to make a hierarchical cluster tree using
average distance between cluster elements:
z1=linkage(d','average');
- For more options in constructing the cluster tree, type help linkage at the Matlab command line, also see a modeling text such as Leach [10].
- We display the dendrogram of the clusters in z1:
h101=figure(101);
dendrogram(z1);
- And, finally, paste in some magic to place the labels correctly:
hx=get(get(h101,'CurrentAxes'),'XTickLabel');
for i=1:size(hx,1)
hx(i)=str2double(hx(i));
end
set(get(h101,'CurrentAxes'),'XTickLabel',[l(hx(1)), ...
l(hx(2)),l(hx(3)),l(hx(4)),l(hx(5)),l(hx(6)),l(hx(7))])
figure(h101);
title('Molecular Phylogenetic Tree');
xlabel('Protein (pdb code)')
ylabel('1-Similarity (%)')
- Print out the dendrogram, or copy it down on paper, along with the names of
the proteins. Refer to Table 6 to write, under each name, the
domain of life each protein originates from.
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...hat we know about the evolution of the three domains of life?}
\end{minipage} }](img229.gif)


Next: Other bioinformatics tools
Up: Bioinformatics Tutorial
Previous: Viewing conserved domains of
zan@uiuc.edu
![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...hat we know about the evolution of the three domains of life?}
\end{minipage} }](img229.gif)