



Next: Geometric path collective variables
Up: Defining collective variables
Previous: Protein structure descriptors
Contents
Index
Subsections
The cartesian {...} block defines a component returning a flat vector containing
the Cartesian coordinates of all participating atoms, in the order
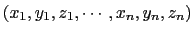 .
.
List of keywords (see also ![[*]](crossref.png) for additional options):
for additional options):
-
atoms
 Group of atoms
Group of atoms
Context: cartesian
Acceptable values: Block atoms {...}
Description: Defines the atoms whose coordinates make up the value of the component.
If rotateReference or centerReference are defined, coordinates
are evaluated within the moving frame of reference.
The distancePairs {...} block defines a
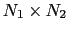 -dimensional variable that includes all mutual distances between the atoms of two groups.
This can be useful, for example, to develop a new variable defined over two groups, by using the scriptedFunction feature.
-dimensional variable that includes all mutual distances between the atoms of two groups.
This can be useful, for example, to develop a new variable defined over two groups, by using the scriptedFunction feature.
List of keywords (see also ![[*]](crossref.png) for additional options):
for additional options):
-
group1: see definition of group1 (distance component)
-
group2: analogous to group1
-
forceNoPBC: see definition of forceNoPBC (distance component)
This component returns a
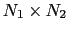 -dimensional vector of numbers, each ranging from 0
to the largest possible distance within the chosen boundary conditions.
-dimensional vector of numbers, each ranging from 0
to the largest possible distance within the chosen boundary conditions.




Next: Geometric path collective variables
Up: Defining collective variables
Previous: Protein structure descriptors
Contents
Index
vmd@ks.uiuc.edu
![]() .
.
![[*]](crossref.png) for additional options):
for additional options):
![]() -dimensional variable that includes all mutual distances between the atoms of two groups.
This can be useful, for example, to develop a new variable defined over two groups, by using the scriptedFunction feature.
-dimensional variable that includes all mutual distances between the atoms of two groups.
This can be useful, for example, to develop a new variable defined over two groups, by using the scriptedFunction feature.
![[*]](crossref.png) for additional options):
for additional options):