



Next: Hot Keys
Up: Using the Mouse in
Previous: Mouse Modes
Contents
Index
Subsections
Pick Item Mode
The mouse can also be used to select things from the screen. As
with many molecular graphics programs, an atom can
be picked by moving the cursor over it and clicking the left mouse
button. When an atom is picked for the first time, a text label
appears which shows the atom residue name and number, and the atom
name. Clicking on the atom again turns the label off.
Picking atoms with the mouse is used to turn on or off various types
of labels, to query for information about an object, or to move items
around on the screen. You can label an atom (and display the atom
name), or you can label geometric values such as the
distance between two atoms (a bond label), an angle between three
atoms (an angle label), or the dihedral angle formed by four
atoms (a dihedral label). This is done by setting the mouse
into the proper picking mode and then selecting the relevant atoms
with the mouse.
You first select the proper picking mode by using the Mouse form, and
choosing the mode required to perform the desired action.
The available actions when in pick item mode are:
- Pick Item Mode
 Query
Query
Clicking with the left or middle mouse button on an item
will print out the name of the item (e.g. the atom name) to the text console
window.
The hot key to set this mode is `0'.
- Pick Item Mode
 Center
Center
This mode is used to change the point about which a molecule
rotates when the molecule is rotated. To cause a molecule to rotate
about a specific atom, select this mode and then click on that atom.
The rotation point may be restored to its default position (the center
of volume of the molecule) by executing the `Reset View' option from
the Mouse form. The hot key to set the mouse to Pick Item
Mode  Centering mode is `c'.
Centering mode is `c'.
- Pick Item Mode
 Atom
Atom
Clicking on an atom will toggle on/off a label for the atom. The hot
key to set this mode is `1'.
- Pick Item Mode
 Bond
Bond
Clicking on two atoms in a row will toggle on/off a bond distance
label between the two atoms (a dotted line with the distance printed
at the midpoint). The hot key to set this mode is `2'.
- Pick Item Mode
 Angle
Angle
Clicking on three atoms in a row will toggle on/off a label showing
the angle formed by the three atoms. The hot key to set this mode is
`3'.
- Pick Item Mode
 Dihedral
Dihedral
Clicking on four atoms in a row toggles on/off a label showing the
dihedral angle formed by the four atoms. The hot key to set this mode
is `4'.
- Pick Item Mode
 MoveAtom
MoveAtom
In this mode, the position of an atom can be changed by clicking on
the desired atom, and dragging with the mouse while the button is still
pressed. This will change the atom coordinates.
The hot key to set this mode is `5'.
- Pick Item Mode
 MoveResidue
MoveResidue
This mode may be used to move all the atoms in a selected residue at
the same time. Select an atom in a residue, and move it to a new
position while keeping the mouse button pressed. All the atoms in the
same residue as the selected one will be moved the same amount.
Holding down the <shift> key and the left mouse button while moving the mouse
will rotate the atoms in the residue about the selected atom. If the middle
mouse button is held down instead, the atoms in the residue will rotate about
a line drawn through the picked atom and parallel to a line coming directly
out of the screen. This behavior is similar to the usual Rotate mode, except
that coordinates of atoms are changed.
The hot key to set this mode is `6'.
- Pick Item Mode
 MoveFragment
MoveFragment
A fragment is a set of atoms all connected by a series of
covalent bonds. This mode acts just like MoveResidue, except that the
atoms which are moved are all in the selected fragment rather than in
the selected residue. This will change the atom coordinates.
Holding down the <shift> key and the left mouse button while moving the mouse
will rotate the atoms in the fragment about the selected atom. If the middle
mouse button is held down instead, the atoms in the fragment will rotate about
a line drawn through the picked atom and parallel to a line coming directly
out of the screen. This behavior is similar to the usual Rotate mode, except
that coordinates of atoms are changed.
The hot key to set this mode is `7'.
- Pick Item Mode
 MoveMolecule
MoveMolecule
This mode may be used to move all the atoms in a selected molecule at
the same time. Select an atom in a molecule, and move it to a new
position while keeping the mouse button pressed. All the atoms in the
same molecule as the selected one will be moved the same amount.
Holding down the <shift> key and the left mouse button while moving the mouse
will rotate the atoms in the molecule about the selected atom. If the middle
mouse button is held down instead, the atoms in the fragment will rotate about
a line drawn through the picked atom and parallel to a line coming directly
out of the screen. This behavior is similar to the usual Rotate mode, except
that coordinates of atoms are changed.
The hot key to set this mode is `8'.
When a label is added to a molecule (say, for a bond, or just to show
the name of an atom), a new entry will be added to the list of current
labels which is available via the
Labels form.
These controls also contain
options to turn the labels on or off, or to delete them entirely from memory.
Fit Submenu
When one has two similar structures, one often wants to compare them.
What's the difference between two X-ray structures? How much did the
structure change during a simulation? To answer these questions, you
must first figure out how to compare two structures, which usually
means that you must find the root mean square deviation (RMSD).
Formally, given 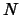 atom positions from structure
atom positions from structure 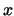 and the
corresponding
and the
corresponding 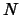 atoms from structure
atoms from structure 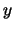 with a weighting factor
with a weighting factor
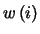 ,
the RMSD is defined as:
,
the RMSD is defined as:
Using this equation by itself probably won't give you the answer you
are looking for. Imagine two identical structures offset by some
distance. The RMSD should be 0, but the offset prevents that from
happening. What you really want is the minimum RMSD between two given
structures; the best fit. There are many ways to do this, but for VMD
we have implemented the method of Kabsch (Acta Cryst. (1978) A34,
827-828 or see file Measure.C in the VMD source code).
This algorithm computes the transformation, needed to move one structure
onto another in order to minimize the RMSD.
With the mathematical prerequisites behind us, we still need to be
able to specify how to choose the atoms to compare. If you want to
compare all the atoms in both structures, and they both have the same
number of atoms, then the problem is easy - 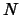 is everything. This
occurs most often in MD simulations when the only thing different
between two structures are the coordinates.
is everything. This
occurs most often in MD simulations when the only thing different
between two structures are the coordinates.
But what about homologous sequences? In this case, the number of
atoms differ because while the number of residues is the same, the
sidechains have different numbers of atoms. The usual solution is to
determine the RMSD based solely on the backbone atoms or, in some
X-ray structures where only the 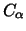 atoms have been
determined, based on the
atoms have been
determined, based on the 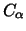 atoms. In addition, VMD allows two other
methods for fitting. Fitting by heavy atoms omits the hydrogens,
since their positions are often not well determined. Fitting by
``picked atoms'' performs only a translation to bring one atom directly
on top of another molecule.
atoms. In addition, VMD allows two other
methods for fitting. Fitting by heavy atoms omits the hydrogens,
since their positions are often not well determined. Fitting by
``picked atoms'' performs only a translation to bring one atom directly
on top of another molecule.
Hopefully the previous discussion revealed the importance of the
options available in the fit submenu. Before examining each of them
in turn, you should be aware of some VMD definitions. In VMD, a
``molecule'' refers to all the atoms from a structure file. The file may
contain multiple molecules under standard chemical usage, but VMD
still thinks of them as one molecule. Instead, those individual parts
are called ``fragments,'' for lack of a better term. With this said,
the fit options in the submenu include:
- Fit
 Do RMSD Fit
Do RMSD Fit
The fitting will change the stored coordinates of one of the molucules.
- Fit
 Print RMSD
Print RMSD
VMD wll print out the RMSD between the molecules after fitting them together,
without changing their stored coordinates.
- Fit
 Two Molecules
Two Molecules
- Fit
 Two Fragments
Two Fragments
These two submenus contain the same options, which select which atoms of
the two molecules should be used to do the least squares fit. For the first
submenu, the atoms used will be taken from all the atoms in two different
molecules; for the second submenu, the atoms used will only be taken from
two separate fragments, where a fragment is a connected chain of residues.
The available atom selections for fitting the molecules are:
- All atoms;
- Heavy atoms (i.e., all except hydrogen);
- Backbone atoms;
- CA (that is, C
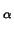 ) atoms;
) atoms;
- Picked atoms (i.e., atoms currently labeled on the screen).
Presently the VMD feedback for RMSD picks is a little terse. To make
things clear, here is the process: First select one of the options
(i.e., All atoms; Heavy atoms; etc.) from the fit submenu. VMD will
respond with a confirmation of your choice and then it will ask you to
click on one atom of each of the two selections you would like to
compare. The selection you click on first will be moved to the
selection you click on second. The atoms that you click on are
representative of the entire molecule, fragment, set of heavy atoms,
etc. associated with those atoms. Thus, if you have chosen to RMSD
fit two molecules, then clicking on one atom for each of the two
molecules does in fact specify an RMSD calculation involving
all atoms of the two molecules.
As a simple example of computing RMSD, load the same molecule twice.
Press the `8' key (this puts VMD into the ``MoveMolecule'' pick mode
and lets you use the mouse to change the coordinates of a molecule).
Click and drag one of the molecules away from the other so there is a
space between the two.
By default the fit routines are configured to do the best fit but for
this example you will only compute the RMSD, so pick ``Fit
 Print RMSD.'' Now go to the Mouse form and pick ``Fit
Print RMSD.'' Now go to the Mouse form and pick ``Fit
 Two Molecules
Two Molecules  All Atoms.'' Look at the
VMD console window and you'll see a new line was printed to show the
current state. The mouse should look like a crosshair. Pick one of
the atoms in the first molecule and then one of the atoms in the second
molecule. The value of the RMSD will be printed to the screen, for
example:
All Atoms.'' Look at the
VMD console window and you'll see a new line was printed to show the
current state. The mouse should look like a crosshair. Pick one of
the atoms in the first molecule and then one of the atoms in the second
molecule. The value of the RMSD will be printed to the screen, for
example:
RMSD between the two molecules is: 1.123410
In case of two identical molecules the RMSD calculation should, of course,
give zero as a result.
To stop computing RMSDs, simply change the mouse mode to what
you want to do next, for example, press r to go to the rotate mode.
A discussion of advanced RMSD features available through the scripting
interface can be found in section §.
VMD's atom alignment should be able to handle most common tasks, but
there are some it cannot do. If this occurs, you might want to look
at Andrew Martin's ProFit at
http://www.biochem.ucl.ac.uk/~martin/text/ProFit.readme




Next: Hot Keys
Up: Using the Mouse in
Previous: Mouse Modes
Contents
Index
vmd@ks.uiuc.edu
![\(
RMSD\left(N; x,y\right) = {\left[\frac {\sum_{i=1}^N
w_i {\parallel x_i - y_i \parallel}^2}
{N \sum_{i=1}^N w_i}\right]}^{\frac {1}{2}}
\)](img12.gif)