From: Ishaan Roy (royi_at_uchicago.edu)
Date: Tue Aug 24 2021 - 17:23:39 CDT
Hello,
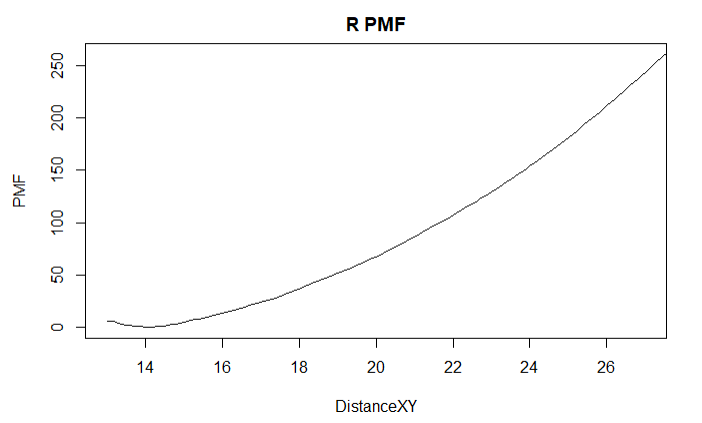
I am using eABF to separate two membrane proteins to find the free binding energy. The PMF values produced in my runs seem to be incredibly high compared to consistently reaching the 100s order of magnitude. We havenít completed a run that plateaus in free energy yet, but this is an example showing what the PMF looks like.
[cid:image001.png_at_01D79905.BA65D100]
A maximum PMF this high already produces a value for the binding constant that is on an order beyond 10^100. Besides being high, it might be worth mentioning that the nonbonded energies are very close to 0 near the end of this run, but the PMF gradient doesnít level off at all. Any ideas on what might be causing this? Or is such a value plausible? This only seems to be a problem with the separation runs; my other colvars produce PMF plots that are closer to what Iíve seen.
Thanks,
Ishaan

This archive was generated by hypermail 2.1.6 : Fri Dec 31 2021 - 23:17:11 CST