From: Susmita Ghosh (g.susmita6_at_gmail.com)
Date: Mon Nov 28 2016 - 23:50:26 CST
Hi all,
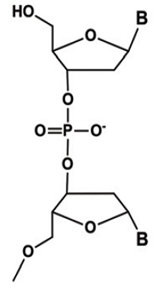
In charmm topology file for a DNA, in general residues are connected
through 3'-5' phosphodiester bond. I want to add extra nucleic acid residue
to a DNA sequence through a 3'-3' phophodister linkage i.e. the phosphate
backbone of extra residue will be connected to 3' end and I have to apply
a patch to 5'-end by adding a hydrogen atom to 05' atom which is different
from the 5TER patch. To change the topology file for the extra residue many
information about the group atoms, bonds, angles and dihedrals need to be
changed which is seemed to be difficult to me. I have tried it by topotools
of vmd by adding a bond between 03'-P and breaking bond between O5'-P. But
this idea is not working as there is no such patching option for 05' atom
in topotools. Could anyone please suggest me how to create 3'-3'
phosphodiester bond for NAMD simulation? The picture for the 3'-3'
phosphodister bond is shown below:
[image: Inline image 1]
Susmita Ghosh
Research Scholar,
Department of Phyics, IIT Guwahati.
Assam, India.

This archive was generated by hypermail 2.1.6 : Sun Dec 31 2017 - 23:20:50 CST