From: jnsong (jnsong_at_itcs.ecnu.edu.cn)
Date: Thu Dec 16 2010 - 07:40:38 CST
Dear all,
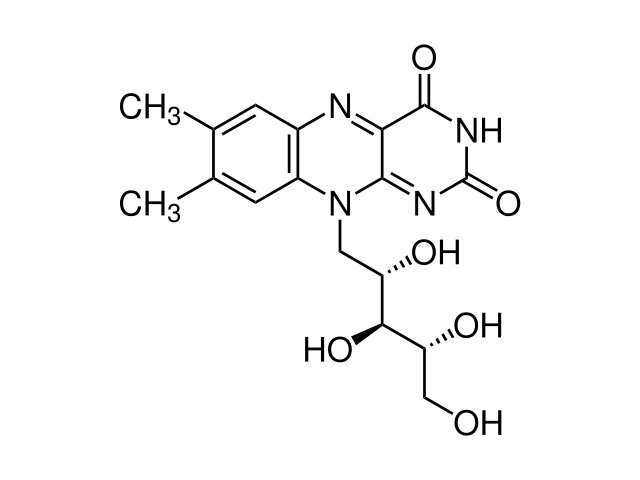
Recently, I want to do MD simulation on a protein and ligand compound
using NAMD with CHARMM force filed.
But I have no idea about how to get topology and parameter file for
the ligand.
Would you please give me some suggestions?
Thank you very much!
Attachment is the ligand I want to get topology and parameter file.
Jianing

This archive was generated by hypermail 2.1.6 : Wed Feb 29 2012 - 15:54:52 CST