



Next: AMBER file and force
Up: Input and Output Files
Previous: File formats
Contents
Index
Subsections
NAMD configuration parameters
- coordinates
 coordinate PDB file
coordinate PDB file 
Acceptable Values: UNIX filename
Description: The PDB file containing initial position coordinate data.
Note that path names can be either absolute or relative.
Only one value may be specified.
- structure
 PSF file
PSF file 
Acceptable Values: UNIX filename
Description: The X-PLOR format PSF file describing the molecular
system to be simulated.
Only one value may be specified.
- parameters
 parameter file
parameter file 
Acceptable Values: UNIX filename
Description: A CHARMM19, CHARMM22, or CHARMM27 parameter file that defines all or part
of the parameters necessary for the molecular system to be simulated.
At least one parameter file must be specified for each simulation.
Multiple definitions (but only one file per definition)
are allowed for systems that require more than one parameter file.
The files will be read
in the order that they appear in the configuration file. If duplicate
parameters are read, a warning message is printed and the last
parameter value read is used. Thus, the order that files are read
can be important in cases where duplicate values appear in
separate files.
- paraTypeXplor
 Is the parameter file in X-PLOR format?
Is the parameter file in X-PLOR format? 
Acceptable Values: on or off
Default Value: on
Description: Specifies whether or not the parameter file(s) are in X-PLOR format.
X-PLOR format is the default for parameter files!
Caveat: The PSF file should be also constructed with X-PLOR in
case of an X-PLOR parameter file because X-PLOR stores information
about the multiplicity of dihedrals in the PSF file. See the X-PLOR
manual for details.
- paraTypeCharmm
 Is the parameter file in CHARMM format?
Is the parameter file in CHARMM format? 
Acceptable Values: on or off
Default Value: off
Description: Specifies whether or not the parameter file(s) are in CHARMM format.
X-PLOR format is the default for parameter files!
Caveat: The information about multiplicity of dihedrals will be
obtained directly from the parameter file, and the full multiplicity
will be used (same behavior as in CHARMM). If the PSF file originates
from X-PLOR, consecutive multiple entries for the same dihedral
(indicating the dihedral multiplicity for X-PLOR) will be ignored.
- velocities
 velocity PDB file
velocity PDB file 
Acceptable Values: UNIX filename
Description: The PDB file containing the initial velocities for all
atoms in the simulation.
This is typically a restart file or final velocity file written
by NAMD during a previous simulation.
Either the temperature
or the velocities/binvelocities
option must be defined to determine an initial set of velocities.
Both options cannot be used together.
- binvelocities
 binary velocity file
binary velocity file 
Acceptable Values: UNIX filename
Description: The binary file containing initial velocities for all
atoms in the simulation.
A binary velocity file is created as output from NAMD
by activating the binaryrestart or binaryoutput options.
The binvelocities option should be used as
an alternative to velocities.
Either the temperature
or the velocities/binvelocities
option must be defined to determine an initial set of velocities.
Both options cannot be used together.
- bincoordinates
 binary coordinate restart file
binary coordinate restart file 
Acceptable Values: UNIX filename
Description:
The binary restart file containing initial position
coordinate data.
A binary coordinate restart file is created as output from NAMD
by activating the binaryrestart or binaryoutput options.
Note that, in the current implementation at least,
the bincoordinates option must be used in addition
to the coordinates option,
but the positions specified by coordinates will then be ignored.
- cwd
 default directory
default directory 
Acceptable Values: UNIX directory name
Description: The default directory for input and output files.
If a value is given, all filenames that
do not begin with a / are assumed to be in this directory.
For example, if cwd is set to /scr, then a
filename of outfile would be modified to /scr/outfile
while a filename of /tmp/outfile would remain unchanged.
If no value for cwd is specified, than all filenames are
left unchanged but are assumed to be relative to the directory
which contains the configuration file given on the command line.
- outputname
 output file prefix
output file prefix 
Acceptable Values: UNIX filename prefix
Description: At the end of every simulation, NAMD writes two files, one
containing the final coordinates and another containing
the final velocities of all atoms in the simulation.
This option specifies the file prefix for these two files as
well as the default prefix for trajectory and restart files.
The position coordinates will be saved to a file named as this prefix
with .coor appended.
The velocities will be saved to a file
named as this prefix with .vel appended.
For example,
if the prefix specified using this option was /tmp/output,
then the two files
would be /tmp/output.coor and /tmp/output.vel.
- binaryoutput
 use binary output files?
use binary output files? 
Acceptable Values: yes or no
Default Value: yes
Description:
Enables the use of binary output files.
If this option is not set to no, then the final output files
will be written in binary rather than PDB format.
Binary files preserve more accuracy between NAMD restarts
than ASCII PDB files,
but the binary files are not guaranteed to be transportable
between computer architectures. (The atom count record is used
to detect wrong-endian files, which works for most atom counts.
The utility program flipbinpdb is provided
to reformat these files if necessary.)
- restartname
 restart files prefix
restart files prefix 
Acceptable Values: UNIX filename prefix
Default Value: outputname.restart
Description:
The prefix to use for restart filenames.
NAMD produces restart files
that store the current positions and velocities of all
atoms at some step of the simulation.
This option specifies the prefix to use for restart
files in the same way that outputname
specifies a filename prefix for the final
positions and velocities.
If restartname is defined then
the parameter restartfreq must also be defined.
- restartfreq
 frequency of restart file generation
frequency of restart file generation 
Acceptable Values: positive integer
Description:
The number of timesteps between the generation of restart files.
- restartsave
 use timestep in restart filenames?
use timestep in restart filenames? 
Acceptable Values: yes or no
Default Value: no
Description:
Appends the current timestep to the restart filename prefix, producing
a sequence of restart files rather than only the last version written.
- binaryrestart
 use binary restart files?
use binary restart files? 
Acceptable Values: yes or no
Default Value: yes
Description:
Enables the use of binary restart files.
If this option is not set to no, then the restart files
will be written in binary rather than PDB format.
Binary files preserve more accuracy between NAMD restarts
than ASCII PDB files,
but the binary files are not guaranteed to be transportable
between computer architectures. (The atom count record is used
to detect wrong-endian files, which works for most atom counts.
The utility program flipbinpdb is provided
to reformat these files if necessary.)
- DCDfile
 coordinate trajectory output file
coordinate trajectory output file 
Acceptable Values: UNIX filename
Default Value: outputname.dcd
Description:
The binary DCD position coordinate trajectory filename.
This file stores the trajectory of all atom position coordinates
using the same format (binary DCD) as X-PLOR.
If DCDfile is defined, then DCDfreq must also be defined.
- DCDfreq
 timesteps between writing coordinates to trajectory file
timesteps between writing coordinates to trajectory file 
Acceptable Values: positive integer
Description:
The number of timesteps between the writing of position coordinates
to the trajectory file.
The initial positions will not be included in the trajectory file.
Positions in DCD files are stored in Å.
- DCDUnitCell
 write unit cell data to dcd file?
write unit cell data to dcd file? 
Acceptable Values: yes or no
Default Value: yes if periodic cell
Description:
If this option is set to yes, then DCD files will contain unit
cell information in the style of Charmm DCD files.
By default this option is enabled if the simulation cell is periodic
in all three dimensions and disabled otherwise.
- velDCDfile
 velocity trajectory output file
velocity trajectory output file 
Acceptable Values: UNIX filename
Default Value: outputname.veldcd
Description:
The binary DCD velocity trajectory filename.
This file stores the trajectory of
all atom velocities using the same format (binary DCD) as X-PLOR.
If velDCDfile is defined, then velDCDfreq must also
be defined.
- velDCDfreq
 timesteps between writing velocities to trajectory file
timesteps between writing velocities to trajectory file 
Acceptable Values: positive integer
Description:
The number of timesteps between the writing of
velocities to the trajectory file.
The initial velocities will not be included in the trajectory file.
Velocities in DCD files are stored in NAMD internal
units and must be multiplied by PDBVELFACTOR=20.45482706
to convert to Å/ps.
- forceDCDfile
 force trajectory output file
force trajectory output file 
Acceptable Values: UNIX filename
Default Value: outputname.forcedcd
Description:
The binary DCD force trajectory filename.
This file stores the trajectory of
all atom forces using the same format (binary DCD) as X-PLOR.
If forceDCDfile is defined, then forceDCDfreq must also
be defined.
- forceDCDfreq
 timesteps between writing force to trajectory file
timesteps between writing force to trajectory file 
Acceptable Values: positive integer
Description:
The number of timesteps between the writing of
forces to the trajectory file.
The initial forces will not be included in the trajectory file.
Forces in DCD files are stored in kcal/mol/Å.
In the current implementation only those forces that are
evaluated during the timestep that a frame is written are
included in that frame. This is different from the behavior
of TclForces and is likely to change based on user feedback.
For this reason it is strongly recommended that forceDCDfreq
be a multiple of fullElectFrequency.
- crashOutputFlag
 bitmask controling the output of atom positions and
velocities when crashing (only used in the GPU-resident mode)
bitmask controling the output of atom positions and
velocities when crashing (only used in the GPU-resident mode)

Acceptable Values: positive integer
Default Value: 1
Description:
The value of crashOutputFlag is used as a bitmask to determine
if CSV files are saved with atom positions and velocities when
crashing. Setting it to 1 will make NAMD write the CSV files
when atoms are moving too fast.
- crashFile
 The CSV filename prefix for saving atom positions and velocities
when NAMD crashes
The CSV filename prefix for saving atom positions and velocities
when NAMD crashes 
Acceptable Values: UNIX filename
Default Value: outputname.crash.csv
Description:
The CSV filename for saving atom positions and velocities when NAMD crashes.
The CSV file contains 7 columns containing the atom IDs in the PDB indexing,
positions and velocities.
NAMD logs a variety of summary information to standard output.
The standard units used by NAMD are
Angstroms for length, kcal/mol for energy,
Kelvin for temperature, and bar for pressure.
Wallclock or CPU times are given in seconds unless otherwise noted.
BOUNDARY energy is from spherical boundary conditions and harmonic restraints,
while MISC energy is from external electric fields and various steering forces.
TOTAL is the sum of the various potential energies, and the KINETIC energy.
TOTAL2 uses a slightly different kinetic energy that is better conserved
during equilibration in a constant energy ensemble.
Although TOTAL2 is calculated internally for producing TOTAL3,
its value is no longer output during simulation.
TOTAL3 is another variation with much smaller short-time fluctuations that
is also adjusted to have the same running average as TOTAL2.
Defects in constant energy simulations are much easier to spot in TOTAL3
than in TOTAL or TOTAL2.
PRESSURE is the pressure calculated based on individual atoms, while
GPRESSURE incorporates hydrogen atoms into the heavier atoms to which
they are bonded, producing smaller fluctuations.
The TEMPAVG, PRESSAVG, and GPRESSAVG are the average of temperature and
pressure values since the previous ENERGY output; for the first step
in the simulation they will be identical to TEMP, PRESSURE, and GPRESSURE.
Performance for NAMD's new GPU-resident mode is improved by avoiding
calculation of energy and virial reductions except when needed.
This means that the averages are no longer calculated over all values
since the previous ENERGY output. Instead, a moving average is
calculated over the previously output values, using a fixed window size
defined by a config file parameter.
GPU-resident mode replaces TOTAL3 with TOTALAVG,
a moving average over the previous TOTAL energy values.
- outputEnergies
 timesteps between energy output
timesteps between energy output 
Acceptable Values: positive integer
Default Value: 1
Description:
The number of timesteps between each energy output of NAMD.
This value
specifies how often NAMD should output the current energy
values to stdout (which can be redirected to a file).
By default, this is done every step.
For long simulations,
the amount of output generated by NAMD can be greatly reduced
by outputting the energies only occasionally.
For GPU-resident mode, the default value is raised to 100,
although performance might be improved by setting it higher.
- movingAverageWindowSize
 fixed window size for calculating moving averages
fixed window size for calculating moving averages 
Acceptable Values: positive integer
Default Value: 20
Description:
In GPU-resident mode, the energy, temperature, and pressure averages
are calculated as moving averages over the previously outputted values.
Changing the window size does not affect performance.
- computeEnergies
 timesteps between energy evaluation
timesteps between energy evaluation 
Acceptable Values: positive integer
Default Value: outputEnergies
Description:
The number of timesteps between each energy evaluation of NAMD.
This value specifies how often NAMD should compute the current energy
values. The default value is to keep the same as outputEnergies.
Other operations requiring the energies, involving outputEnergies,
alchOutFreq and the period of calling a callback TCL procedure,
should be a multiple of this value.
In the GPU build of NAMD, increasing the value of this option can
improve the performance since the evaluation of energies requires
double-precision mathematical operations.
- outputEnergiesPrecision
 energy output precision
energy output precision 
Acceptable Values: positive integer
Default Value: 4
Description:
The number of decimal digits used when printing NAMD energies.
Increasing the output precision will skew the aligned 80-column format
that NAMD has traditionally provided. To keep energy output aligned,
increase the terminal width by 5 columns for each additional decimal digit
beyond 4.
- mergeCrossterms
 add crossterm energy to dihedral?
add crossterm energy to dihedral? 
Acceptable Values: yes or no
Default Value: yes
Description:
If crossterm (or CMAP) terms are present in the potential,
the energy is added to the dihedral energy to avoid altering
the energy output format.
Disable this feature to add a separate ``CROSS'' field to the output.
- outputMomenta
 timesteps between momentum output
timesteps between momentum output 
Acceptable Values: nonnegative integer
Default Value: 0
Description:
The number of timesteps between each momentum output of NAMD.
If specified and nonzero, linear and angular momenta will be
output to stdout.
- outputPressure
 timesteps between pressure output
timesteps between pressure output 
Acceptable Values: nonnegative integer
Default Value: 0
Description:
The number of timesteps between each pressure output of NAMD.
If specified and nonzero, atomic and group pressure tensors
will be output to stdout.
- outputTiming
 timesteps between timing output
timesteps between timing output 
Acceptable Values: nonnegative integer
Default Value: the greater of firstLdbStep or 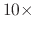 outputEnergies
outputEnergies
Description:
The number of timesteps between each timing output of NAMD.
If nonzero, CPU and wallclock times and memory usage will be
output to stdout.
These data are from node 0 only; CPU times and memory usage for other nodes
may vary.
- outputPerformance
 Print performance statistics?
Print performance statistics? 
Acceptable Values: ``on'' or ``off''
Default Value: on
Description:
When enabled, every ``TIMING:'' line is immediately followed by a
``PERFORMANCE:'' line that states the average ns/day performance.
For the cases when it is preferable to have trajectory output of
a subset of the atoms, selections can be defined and assigned
independent output frequencies and output file names.
Atom selections are defined in one of two ways.
The first method is by creating a PDB file where the temperaturefactor field (AKA beta in VMD) is assigned a
value greater than zero for each atom in the selection. For example, when the objective is to have a more frequent output trajectory containing no solvent, the following VMD commands will produce an appropriate selection PDB.
(Note: example assumes structure and coordinates are already loaded)
set allatoms [atomselect top "all"]
set soluteatoms [atomselect top "not water and not ions"]
$allatoms set beta 0
$soluteatoms set beta 1
$allatoms writepdb solute.pdb
$soluteatoms writepsf solute.psf
The additional PSF file in the prior example is not necessary for NAMD, but would be useful in VMD, or other subsequent analysis, to examine the trajectory associated with given selection of atoms.
The second method is by creating a binary index file containing the binary formatted atom indices within the selection, preceded by a short version field and the total count of atom indices. The VMD indexfile plugin is recommended for this task. See VMD tcl code below. Note, NAMD supports either endian choice for the file, but it must be consistent within the file.
(Note: example assumes structure and coordinates are already loaded)
set soluteatoms [atomselect top "not water and not ions"]
#export via the VMD plugin
package require indexfile
indexfile::write_indexfile $soluteatoms solute.idx
#or use custom tcl code
set myList [$soluteatoms get index]
set $fh [open "solute.idx" wb]
set idxversion 1
set size [$soluteatoms num]
puts -nonewline $fh [binary format s $idxversion]
puts -nonewline $fh [binary format i $size]
foreach num $myList
{
puts -nonewline $fh [binary format i $num]
}
close $fh
Such selections can be used within NAMD using the arguments described
below. A maximum of 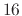 such selections is allowed. There are no
restrictions regarding atoms belonging to multiple selections. Each
selection is identified by a user defined string tag to bind the frequency and input file and output file definitions
together for each selection. The tag is an alphanumeric string
without spaces, up to
such selections is allowed. There are no
restrictions regarding atoms belonging to multiple selections. Each
selection is identified by a user defined string tag to bind the frequency and input file and output file definitions
together for each selection. The tag is an alphanumeric string
without spaces, up to 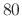 characters in length, which identifies to
which selection the specified field applies.
characters in length, which identifies to
which selection the specified field applies.
In the case of the example above, the following configuration would be valid:
dcdSelection yes
dcdSelectionInputfile soluteonly solute.pdb
dcdSelectionfreq soluteonly 100
dcdSelectionfile soluteonly solute.dcd
NOTE: Using this feature with special memory-optimized NAMD builds requires the binary index file form of selection instead of using a PDB.
https://www.ks.uiuc.edu/Research/namd/wiki/index.cgi?NamdMemoryReduction.
See lib/replica/examples/mplexjob0-dcdselect.conf for replica usage.




Next: AMBER file and force
Up: Input and Output Files
Previous: File formats
Contents
Index
http://www.ks.uiuc.edu/Research/namd/