


Next: About this document ...
Up: NAMD 2.10b1 User's Guide
Previous: Bibliography
Contents
- 1-4scaling parameter
- Non-bonded force field parameters
- name parameter
- Biasing and analysis methods
- colvars parameter
- Biasing and analysis methods
- outputEnergy parameter
- Biasing and analysis methods
- hillWeight parameter
- Metadynamics
- newHillFrequency parameter
- Metadynamics
- writeFreeEnergyFile parameter
- Output files
- saveFreeEnergyFile parameter
- Output files
- useGrids parameter
- Performance tuning
- hillWidth parameter
- Performance tuning
- rebinGrids parameter
- Performance tuning
- wellTempered parameter
- Well-tempered metadynamics
- biasTemperature parameter
- Well-tempered metadynamics
- multipleReplicas parameter
- Multiple-replicas metadynamics
- replicaID parameter
- Multiple-replicas metadynamics
- replicasRegistry parameter
- Multiple-replicas metadynamics
- replicaUpdateFrequency parameter
- Multiple-replicas metadynamics
- dumpPartialFreeEnergyFile parameter
- Multiple-replicas metadynamics
- name parameter
- Compatibility and post-processing
- keepHills parameter
- Compatibility and post-processing
- writeHillsTrajectory parameter
- Compatibility and post-processing
- forceConstant parameter
- Harmonic restraints
- centers parameter
- Harmonic restraints
- targetCenters parameter
- Moving restraints: steered molecular
- targetNumSteps parameter
- Moving restraints: steered molecular
- outputCenters parameter
- Moving restraints: steered molecular
- outputAccumulatedWork parameter
- Moving restraints: steered molecular
- targetNumStages parameter
- Moving restraints: umbrella sampling
- targetForceConstant parameter
- Changing force constant
- targetForceExponent parameter
- Changing force constant
- targetEquilSteps parameter
- Changing force constant
- lambdaSchedule parameter
- Changing force constant
- colvars parameter
- NAMD parameters
- colvarsConfig parameter
- NAMD parameters
- colvarsInput parameter
- NAMD parameters
- colvarsTrajFrequency parameter
- Configuration file for the
- colvarsTrajAppend parameter
- Configuration file for the
- colvarsRestartFrequency parameter
- Configuration file for the
- indexFile parameter
- Configuration file for the
- analysis parameter
- Configuration file for the
- name parameter
- General options for a
- width parameter
- General options for a
- lowerBoundary parameter
- General options for a
- upperBoundary parameter
- General options for a
- hardLowerBoundary parameter
- General options for a
- hardUpperBoundary parameter
- General options for a
- expandBoundaries parameter
- General options for a
- lowerWallConstant parameter
- Artificial boundary potentials (walls)
- lowerWall parameter
- Artificial boundary potentials (walls)
- upperWallConstant parameter
- Artificial boundary potentials (walls)
- upperWall parameter
- Artificial boundary potentials (walls)
- outputValue parameter
- Trajectory output
- outputVelocity parameter
- Trajectory output
- outputEnergy parameter
- Trajectory output
- outputSystemForce parameter
- Trajectory output
- outputAppliedForce parameter
- Trajectory output
- extendedLagrangian parameter
- Extended Lagrangian.
- extendedFluctuation parameter
- Extended Lagrangian.
- extendedTimeConstant parameter
- Extended Lagrangian.
- extendedTemp parameter
- Extended Lagrangian.
- extendedLangevinDamping parameter
- Extended Lagrangian.
- corrFunc parameter
- Statistical analysis of collective
- corrFuncWithColvar parameter
- Statistical analysis of collective
- corrFuncType parameter
- Statistical analysis of collective
- corrFuncNormalize parameter
- Statistical analysis of collective
- corrFuncLength parameter
- Statistical analysis of collective
- corrFuncStride parameter
- Statistical analysis of collective
- corrFuncOffset parameter
- Statistical analysis of collective
- corrFuncOutputFile parameter
- Statistical analysis of collective
- runAve parameter
- Statistical analysis of collective
- runAveLength parameter
- Statistical analysis of collective
- runAveStride parameter
- Statistical analysis of collective
- runAveOutputFile parameter
- Statistical analysis of collective
- atomNumbers parameter
- Selection keywords
- indexGroup parameter
- Selection keywords
- atomNumbersRange parameter
- Selection keywords
- atomNameResidueRange parameter
- Selection keywords
- psfSegID parameter
- Selection keywords
- atomsFile parameter
- Selection keywords
- atomsCol parameter
- Selection keywords
- atomsColValue parameter
- Selection keywords
- dummyAtom parameter
- Selection keywords
- centerReference parameter
- Moving frame of reference.
- rotateReference parameter
- Moving frame of reference.
- refPositions parameter
- Moving frame of reference.
- refPositionsFile parameter
- Moving frame of reference.
- refPositionsCol parameter
- Moving frame of reference.
- refPositionsColValue parameter
- Moving frame of reference.
- refPositionsGroup parameter
- Moving frame of reference.
- enableFitGradients parameter
- Moving frame of reference.
- enableForces parameter
- Moving frame of reference.
- group1 parameter
- distance: center-of-mass distance between
- group2 parameter
- distance: center-of-mass distance between
- forceNoPBC parameter
- distance: center-of-mass distance between
- oneSiteSystemForce parameter
- distance: center-of-mass distance between
- main parameter
- distanceZ: projection of a
- ref parameter
- distanceZ: projection of a
- ref2 parameter
- distanceZ: projection of a
- axis parameter
- distanceZ: projection of a
- forceNoPBC parameter
- distanceZ: projection of a
- oneSiteSystemForce parameter
- distanceZ: projection of a
- exponent parameter
- distanceInv: mean distance between
- oneSiteSystemForce parameter
- dihedral: torsional angle between
- cutoff parameter
- coordNum: coordination number between
- cutoff3 parameter
- coordNum: coordination number between
- expNumer parameter
- coordNum: coordination number between
- expDenom parameter
- coordNum: coordination number between
- group2CenterOnly parameter
- coordNum: coordination number between
- atoms parameter
- rmsd: root mean square
- refPositions parameter
- rmsd: root mean square
- refPositionsFile parameter
- rmsd: root mean square
- refPositionsCol parameter
- rmsd: root mean square
- refPositionsColValue parameter
- rmsd: root mean square
- vector parameter
- eigenvector: projection of the
- vectorFile parameter
- eigenvector: projection of the
- vectorCol parameter
- eigenvector: projection of the
- vectorColValue parameter
- eigenvector: projection of the
- differenceVector parameter
- eigenvector: projection of the
- axis parameter
- inertiaZ: total moment of
- closestToQuaternion parameter
- orientation: orientation from reference
- axis parameter
- spinAngle: angle of rotation
- residueRange parameter
- alpha:
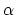 -helix content
-helix content
- psfSegID parameter
- alpha:
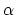 -helix content
-helix content
- hBondCoeff parameter
- alpha:
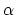 -helix content
-helix content
- angleRef parameter
- alpha:
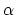 -helix content
-helix content
- angleTol parameter
- alpha:
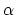 -helix content
-helix content
- hBondCutoff parameter
- alpha:
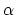 -helix content
-helix content
- hBondExpNumer parameter
- alpha:
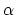 -helix content
-helix content
- hBondExpDenom parameter
- alpha:
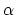 -helix content
-helix content
- vectorFile parameter
- dihedralPC: protein dihedral pricipal
- vectorNumber parameter
- dihedralPC: protein dihedral pricipal
- period parameter
- Periodic components.
- wrapAround parameter
- Periodic components.
- componentCoeff parameter
- Linear and polynomial combinations
- componentExp parameter
- Linear and polynomial combinations
- abort command
- Tcl scripting interface and
- accelMD parameter
- NAMD parameters
- accelMDalpha parameter
- NAMD parameters
- accelMDdihe parameter
- NAMD parameters
- accelMDdual parameter
- NAMD parameters
- accelMDE parameter
- NAMD parameters
- accelMDFirstStep parameter
- NAMD parameters
- accelMDLastStep parameter
- NAMD parameters
- accelMDOutFreq parameter
- NAMD parameters
- accelMDTalpha parameter
- NAMD parameters
- accelMDTE parameter
- NAMD parameters
- adaptTempBins parameter
- NAMD parameters
- adaptTempCgamma parameter
- NAMD parameters
- adaptTempDt parameter
- NAMD parameters
- adaptTempFirstStep parameter
- NAMD parameters
- adaptTempFreq parameter
- NAMD parameters
- adaptTempInFile parameter
- NAMD parameters
- adaptTempLangevin parameter
- NAMD parameters
- adaptTempLastStep parameter
- NAMD parameters
- adaptTempMD parameter
- NAMD parameters
- adaptTempOutFreq parameter
- NAMD parameters
- adaptTempRandom parameter
- NAMD parameters
- adaptTempRescaling parameter
- NAMD parameters
- adaptTempRestartFile parameter
- NAMD parameters
- adaptTempRestartFreq parameter
- NAMD parameters
- adaptTempTmax parameter
- NAMD parameters
- adaptTempTmin parameter
- NAMD parameters
- alch parameter
- Implementation of the free
- alchCol parameter
- Implementation of the free
- alchDecouple parameter
- Implementation of the free
- alchElecLambdaStart parameter
- Implementation of the free
- alchEquilSteps parameter
- Implementation of the free
- alchFile parameter
- Implementation of the free
- alchLambda parameter
- Tcl scripting interface and
| Implementation of the free
- alchLambda2 parameter
- Tcl scripting interface and
| Implementation of the free
- alchOutFile parameter
- Implementation of the free
- alchOutFreq parameter
- Implementation of the free
- alchType parameter
- Implementation of the free
- alchVdwLambdaEnd parameter
- Implementation of the free
- alchVdwShiftCoeff parameter
- Implementation of the free
- alias psfgen command
- List of Commands
| List of Commands
- alphaCutoff parameter
- Configuration Parameters
- amber parameter
- AMBER force field parameters
- ambercoor parameter
- AMBER force field parameters
- applyBias parameter
- Parameters for ABF
- Atoms moving too fast
- Non-bonded interaction distance-testing
- auto psfgen command
- List of Commands
- Bad global exclusion count
- Non-bonded interaction distance-testing
- BerendsenPressure parameter
- Tcl scripting interface and
| Berendsen pressure bath coupling
- BerendsenPressureCompressibility parameter
- Tcl scripting interface and
| Berendsen pressure bath coupling
- BerendsenPressureFreq parameter
- Berendsen pressure bath coupling
- BerendsenPressureRelaxationTime parameter
- Tcl scripting interface and
| Berendsen pressure bath coupling
- BerendsenPressureTarget parameter
- Tcl scripting interface and
| Berendsen pressure bath coupling
- binaryoutput parameter
- Tcl scripting interface and
| Output files
- binaryrestart parameter
- Output files
- bincoordinates parameter
- Input files
- binvelocities parameter
- Input files
- BOUNDARY energy
- Standard output
- callback command
- Tcl scripting interface and
- cellBasisVector1 parameter
- Periodic boundary conditions
- cellBasisVector2 parameter
- Periodic boundary conditions
- cellBasisVector3 parameter
- Periodic boundary conditions
- cellOrigin parameter
- Periodic boundary conditions
- checkpoint command
- Tcl scripting interface and
- COMmotion parameter
- Initialization
- consexp parameter
- Harmonic restraint parameters
- consForceConfig command
- Tcl scripting interface and
| Constant Forces
- consForceFile parameter
- Constant Forces
- consForceScaling parameter
- Tcl scripting interface and
| Constant Forces
- conskcol parameter
- Harmonic restraint parameters
- conskfile parameter
- Harmonic restraint parameters
- consref parameter
- Harmonic restraint parameters
- constantForce parameter
- Tcl scripting interface and
| Constant Forces
- constraints parameter
- Harmonic restraint parameters
- constraintScaling parameter
- Tcl scripting interface and
| Harmonic restraint parameters
- coord psfgen command
- List of Commands
- coordinates parameter
- Input files
- coordpdb psfgen command
- List of Commands
- coorfile command
- Tcl scripting interface and
- cosAngles parameter
- MARTINI Residue-Based Coarse-Grain Forcefield
- cutoff parameter
- Non-bonded force field parameters
- cwd parameter
- Input files
- cylindricalBC parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCAxis parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCCenter parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCexp1 parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCexp2 parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCk1 parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCk2 parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCl1 parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCl2 parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCr1 parameter
- Cylindrical harmonic boundary conditions
- cylindricalBCr2 parameter
- Cylindrical harmonic boundary conditions
- DCDfile parameter
- Tcl scripting interface and
| Output files
- DCDfreq parameter
- Output files
- DCDUnitCell parameter
- Output files
- delatom psfgen command
- List of Commands
- dielectric parameter
- Non-bonded force field parameters
- drude parameter
- Drude force field parameters
- drudeBondConst parameter
- Drude force field parameters
- drudeBondLen parameter
- Drude force field parameters
- drudeDamping parameter
- Drude force field parameters
- drudeNbTholeCut parameter
- Drude force field parameters
- drudeTemp parameter
- Drude force field parameters
- eField parameter
- Tcl scripting interface and
| External Electric Field
- eFieldFreq parameter
- Tcl scripting interface and
- eFieldNormalized parameter
- External Electric Field
- eFieldOn parameter
- External Electric Field
- eFieldPhase parameter
- Tcl scripting interface and
- error message
- Atoms moving too fast
- Non-bonded interaction distance-testing
- Bad global exclusion count
- Non-bonded interaction distance-testing
- exclude parameter
- Non-bonded force field parameters
- ExcludeFromPressure parameter
- Nosé-Hoover Langevin piston pressure
- ExcludeFromPressureCol parameter
- Nosé-Hoover Langevin piston pressure
- ExcludeFromPressureFile parameter
- Nosé-Hoover Langevin piston pressure
- exit command
- Tcl scripting interface and
- extCoordFilename parameter
- External Program Forces
- extendedSystem parameter
- Periodic boundary conditions
- extForceFilename parameter
- External Program Forces
- extForces parameter
- External Program Forces
- extForcesCommand parameter
- External Program Forces
- extraBonds parameter
- Extra bond, angle, and
- extraBondsFile parameter
- Extra bond, angle, and
- FFTWEstimate parameter
- PME parameters
- FFTWUseWisdom parameter
- PME parameters
- FFTWWisdomFile parameter
- PME parameters
- first psfgen command
- List of Commands
- firsttimestep parameter
- Timestep parameters
- fixedAtoms parameter
- Tcl scripting interface and
| Fixed atoms parameters
- fixedAtomsCol parameter
- Fixed atoms parameters
- fixedAtomsFile parameter
- Fixed atoms parameters
- fixedAtomsForces parameter
- Tcl scripting interface and
| Fixed atoms parameters
- forceDCDfile parameter
- Output files
- forceDCDfreq parameter
- Output files
- FullDirect parameter
- Full direct parameters
- fullElectFrequency parameter
- Multiple timestep parameters
- fullSamples parameter
- Parameters for ABF
- GBIS parameter
- Configuration Parameters
- GBISBeta parameter
- Configuration Parameters
- GBISDelta parameter
- Configuration Parameters
- GBISGamma parameter
- Configuration Parameters
- GoCoordinates parameter
- Configuration file modifications
- GoForcesOn parameter
- Configuration file modifications
- GoMethod parameter
- Configuration file modifications
- GoParameters parameter
- Configuration file modifications
- GPRESSAVG
- Standard output
- GPRESSURE
- Standard output
- grocoorfile parameter
- GROMACS force field parameters
| Configuration file modifications
- gromacs parameter
- GROMACS force field parameters
| Configuration file modifications
- GromacsPair parameter
- Configuration file modifications
- grotopfile parameter
- GROMACS force field parameters
| Configuration file modifications
- guesscoord psfgen command
- List of Commands
- hgroupCutoff (Å) parameter
- Non-bonded interaction distance-testing
- hideJacobian parameter
- Parameters for ABF
- historyFreq parameter
- Parameters for ABF
- IMDfreq parameter
- Interactive Molecular Dynamics (IMD)
- IMDignore parameter
- Interactive Molecular Dynamics (IMD)
- IMDon parameter
- Interactive Molecular Dynamics (IMD)
- IMDport parameter
- Interactive Molecular Dynamics (IMD)
- IMDwait parameter
- Interactive Molecular Dynamics (IMD)
- inputPrefix parameter
- Parameters for ABF
- intrinsicRadiusOffset parameter
- Configuration Parameters
- ionConcentration parameter
- Configuration Parameters
- isset command
- Tcl scripting interface and
- istrue command
- Tcl scripting interface and
- langevin parameter
- Langevin dynamics parameters
- langevinCol parameter
- Langevin dynamics parameters
- langevinDamping parameter
- Langevin dynamics parameters
- langevinFile parameter
- Langevin dynamics parameters
- langevinHydrogen parameter
- Langevin dynamics parameters
- LangevinPiston parameter
- Tcl scripting interface and
| Nosé-Hoover Langevin piston pressure
- LangevinPistonDecay parameter
- Tcl scripting interface and
| Nosé-Hoover Langevin piston pressure
- LangevinPistonPeriod parameter
- Tcl scripting interface and
| Nosé-Hoover Langevin piston pressure
- LangevinPistonTarget parameter
- Tcl scripting interface and
| Nosé-Hoover Langevin piston pressure
- LangevinPistonTemp parameter
- Tcl scripting interface and
| Nosé-Hoover Langevin piston pressure
- langevinTemp parameter
- Tcl scripting interface and
| Langevin dynamics parameters
- last psfgen command
- List of Commands
- les parameter
- Simulation
- lesCol parameter
- Simulation
- lesFactor parameter
- Simulation
- lesFile parameter
- Simulation
- lesReduceMass parameter
- Simulation
- lesReduceTemp parameter
- Simulation
- limitdist parameter
- Non-bonded force field parameters
- LJcorrection parameter
- Non-bonded force field parameters
- longSplitting parameter
- Multiple timestep parameters
- loweAndersen parameter
- Lowe-Andersen dynamics parameters
- loweAndersenCutoff parameter
- Lowe-Andersen dynamics parameters
- loweAndersenRate parameter
- Lowe-Andersen dynamics parameters
- loweAndersenTemp parameter
- Lowe-Andersen dynamics parameters
- margin parameter
- Non-bonded interaction distance-testing
- margin violations
- Non-bonded interaction distance-testing
- martiniDielAllow parameter
- MARTINI Residue-Based Coarse-Grain Forcefield
- martiniSwitching parameter
- MARTINI Residue-Based Coarse-Grain Forcefield
- maxForce parameter
- Parameters for ABF
- maximumMove parameter
- Velocity quenching parameters
- measure command
- Tcl scripting interface and
- mergeCrossterms parameter
- Standard output
- mgridforce parameter
- Grid Forces
- mgridforcechargecol parameter
- Grid Forces
- mgridforcecol parameter
- Grid Forces
- mgridforcecont1 parameter
- Grid Forces
- mgridforcecont2 parameter
- Grid Forces
- mgridforcecont3 parameter
- Grid Forces
- mgridforcefile parameter
- Grid Forces
- mgridforcelite parameter
- Grid Forces
- mgridforcepotfile parameter
- Grid Forces
- mgridforcescale parameter
- Grid Forces
- mgridforcevoff parameter
- Grid Forces
- mgridforcevolts parameter
- Grid Forces
- minBabyStep parameter
- Conjugate gradient parameters
- minimization parameter
- Conjugate gradient parameters
- minimize command
- Tcl scripting interface and
- minLineGoal parameter
- Conjugate gradient parameters
- minTinyStep parameter
- Conjugate gradient parameters
- MISC energy
- Standard output
- molly parameter
- Multiple timestep parameters
- mollyIterations parameter
- Multiple timestep parameters
- mollyTolerance parameter
- Multiple timestep parameters
- movingConstraints parameter
- Moving Constraints
- movingConsVel parameter
- Moving Constraints
- MTSAlgorithm parameter
- Multiple timestep parameters
- multiply psfgen command
- List of Commands
- mutate psfgen command
- List of Commands
- nonbondedFreq parameter
- Multiple timestep parameters
- nonbondedScaling parameter
- Tcl scripting interface and
| Non-bonded force field parameters
- numNodes command
- Tcl scripting interface and
- numPes command
- Tcl scripting interface and
- numPhysicalNodes command
- Tcl scripting interface and
- numsteps parameter
- Timestep parameters
- OPLS
- Non-bonded force field parameters
- output command
- Tcl scripting interface and
| Tcl scripting interface and
- output onlyforces command
- Tcl scripting interface and
- output withforces command
- Tcl scripting interface and
- outputEnergies parameter
- Standard output
- outputFreq parameter
- Parameters for ABF
| Multidimensional histograms
- outputMomenta parameter
- Standard output
- outputname parameter
- Output files
- outputPairlists parameter
- Non-bonded interaction distance-testing
- outputPressure parameter
- Standard output
- outputTiming parameter
- Standard output
- pairInteraction parameter
- Pair interaction calculations
- pairInteractionCol parameter
- Pair interaction calculations
- pairInteractionFile parameter
- Pair interaction calculations
- pairInteractionGroup1 parameter
- Pair interaction calculations
- pairInteractionGroup2 parameter
- Pair interaction calculations
- pairInteractionSelf parameter
- Pair interaction calculations
- pairlistdist parameter
- Non-bonded interaction distance-testing
- pairlistGrow parameter
- Non-bonded interaction distance-testing
- pairlistMinProcs parameter
- Non-bonded interaction distance-testing
- pairlistShrink parameter
- Non-bonded interaction distance-testing
- pairlistsPerCycle parameter
- Non-bonded interaction distance-testing
- pairlistTrigger parameter
- Non-bonded interaction distance-testing
- parameters parameter
- Input files
- paraTypeCharmm parameter
- Input files
- paraTypeXplor parameter
- Input files
- parmfile parameter
- AMBER force field parameters
- patch psfgen command
- List of Commands
- pdb psfgen command
- List of Commands
- pdbalias atom psfgen command
- List of Commands
- pdbalias residue psfgen command
- List of Commands
- PME parameter
- PME parameters
- PMEGridSizeX parameter
- PME parameters
- PMEGridSizeY parameter
- PME parameters
- PMEGridSizeZ parameter
- PME parameters
- PMEGridSpacing parameter
- PME parameters
- PMEInterpOrder parameter
- PME parameters
- PMEProcessors parameter
- PME parameters
- PMETolerance parameter
- PME parameters
- PRESSAVG
- Standard output
- pressureProfile parameter
- Pressure profile calculations
- pressureProfileAtomTypes parameter
- Pressure profile calculations
- pressureProfileAtomTypesCol parameter
- Pressure profile calculations
- pressureProfileAtomTypesFile parameter
- Pressure profile calculations
- pressureProfileEwald parameter
- Pressure profile calculations
- pressureProfileEwaldX parameter
- Pressure profile calculations
- pressureProfileEwaldY parameter
- Pressure profile calculations
- pressureProfileEwaldZ parameter
- Pressure profile calculations
- pressureProfileFreq parameter
- Pressure profile calculations
- pressureProfileSlabs parameter
- Pressure profile calculations
- print command
- Tcl scripting interface and
- psfcontext allcaps psfgen command
- List of Commands
- psfcontext create psfgen command
- List of Commands
- psfcontext delete psfgen command
- List of Commands
- psfcontext eval psfgen command
- List of Commands
- psfcontext mixedcase psfgen command
- List of Commands
- psfcontext psfgen command
- List of Commands
- psfcontext reset psfgen command
- List of Commands
- psfcontext stats psfgen command
- List of Commands
- ramd accel parameter
- Random acceleration molecular dynamics
- ramd debugLevel parameter
- Random acceleration molecular dynamics
- ramd firstProtAtom parameter
- Random acceleration molecular dynamics
- ramd firstRamdAtom parameter
- Random acceleration molecular dynamics
- ramd forceOutFreq parameter
- Random acceleration molecular dynamics
- ramd lastProtAtom parameter
- Random acceleration molecular dynamics
- ramd lastRamdAtom parameter
- Random acceleration molecular dynamics
- ramd maxDist parameter
- Random acceleration molecular dynamics
- ramd mdStart parameter
- Random acceleration molecular dynamics
- ramd mdSteps parameter
- Random acceleration molecular dynamics
- ramd ramdSeed parameter
- Random acceleration molecular dynamics
- ramd ramdSteps parameter
- Random acceleration molecular dynamics
- ramd rMinMd parameter
- Random acceleration molecular dynamics
- ramd rMinRamd parameter
- Random acceleration molecular dynamics
- readexclusions parameter
- AMBER force field parameters
- readpsf psfgen command
- List of Commands
- reassignFreq parameter
- Temperature reassignment parameters
- reassignHold parameter
- Temperature reassignment parameters
- reassignIncr parameter
- Temperature reassignment parameters
- reassignTemp parameter
- Tcl scripting interface and
| Temperature reassignment parameters
- regenerate psfgen command
- List of Commands
| List of Commands
- reinitatoms command
- Tcl scripting interface and
- reinitvels command
- Tcl scripting interface and
- reloadCharges command
- Tcl scripting interface and
- replica exchange
- Replica exchange simulations
- rescaleFreq parameter
- Temperature rescaling parameters
- rescaleTemp parameter
- Tcl scripting interface and
| Temperature rescaling parameters
- rescalevels command
- Tcl scripting interface and
- resetpsf psfgen command
- List of Commands
- residue psfgen command
- List of Commands
- restartfreq parameter
- Output files
- restartname parameter
- Tcl scripting interface and
| Output files
- restartsave parameter
- Output files
- rigidBonds parameter
- Bond constraint parameters
- rigidDieOnError parameter
- Bond constraint parameters
- rigidIterations parameter
- Bond constraint parameters
- rigidTolerance parameter
- Bond constraint parameters
- rotConsAxis parameter
- Rotating Constraints
- rotConsPivot parameter
- Rotating Constraints
- rotConstraints parameter
- Rotating Constraints
- rotConsVel parameter
- Rotating Constraints
- run command
- Tcl scripting interface and
- run norepeat command
- Tcl scripting interface and
- SASA parameter
- Configuration Parameters
- scnb parameter
- AMBER force field parameters
- seed parameter
- Initialization
- segment psfgen command
- List of Commands
- selectConstraints parameter
- Harmonic restraint parameters
- selectConstrX parameter
- Harmonic restraint parameters
- selectConstrY parameter
- Harmonic restraint parameters
- selectConstrZ parameter
- Harmonic restraint parameters
- SMD parameter
- Parameters
- SMDDir parameter
- Parameters
- SMDFile parameter
- Parameters
- SMDk parameter
- Parameters
- SMDk2 parameter
- Parameters
- SMDOutputFreq parameter
- Parameters
- SMDVel parameter
- Parameters
- solventDielectric parameter
- Configuration Parameters
- source command
- Tcl scripting interface and
- sphericalBC parameter
- Spherical harmonic boundary conditions
- sphericalBCCenter parameter
- Spherical harmonic boundary conditions
- sphericalBCexp1 parameter
- Spherical harmonic boundary conditions
- sphericalBCexp2 parameter
- Spherical harmonic boundary conditions
- sphericalBCk1 parameter
- Spherical harmonic boundary conditions
- sphericalBCk2 parameter
- Spherical harmonic boundary conditions
- sphericalBCr1 parameter
- Spherical harmonic boundary conditions
- sphericalBCr2 parameter
- Spherical harmonic boundary conditions
- splitPatch parameter
- Non-bonded interaction distance-testing
- staticAtomAssignment parameter
- Configuration file modifications
- stepspercycle parameter
- Non-bonded interaction distance-testing
- StrainRate parameter
- Nosé-Hoover Langevin piston pressure
- structure parameter
- Input files
- surfaceTension parameter
- Configuration Parameters
- SurfaceTensionTarget parameter
- Tcl scripting interface and
| Nosé-Hoover Langevin piston pressure
- switchdist parameter
- Non-bonded force field parameters
- switching parameter
- Non-bonded force field parameters
- symmetryFile parameter
- Symmetry Restraints
- symmetryFirstFullStep parameter
- Symmetry Restraints
- symmetryFirstStep parameter
- Symmetry Restraints
- symmetryk parameter
- Symmetry Restraints
- symmetrykFile parameter
- Symmetry Restraints
- symmetryLastFullStep parameter
- Symmetry Restraints
- symmetryLastStep parameter
- Symmetry Restraints
- symmetryMatrixFile parameter
- Symmetry Restraints
- symmetryRestraints parameter
- Symmetry Restraints
- symmetryScaleForces parameter
- Symmetry Restraints
- tableInterpType parameter
- Tabulated nonbonded interaction parameters
- tabulatedEnergies parameter
- Tabulated nonbonded interaction parameters
- tabulatedEnergiesFile parameter
- Tabulated nonbonded interaction parameters
- tclBC parameter
- Tcl Boundary Forces
- tclBCArgs parameter
- Tcl Boundary Forces
- tclBCScript parameter
- Tcl Boundary Forces
- tclForces parameter
- Tcl Forces and Analysis
- tclForcesScript parameter
- Tcl Forces and Analysis
- tCouple parameter
- Temperature coupling parameters
- tCoupleCol parameter
- Temperature coupling parameters
- tCoupleFile parameter
- Temperature coupling parameters
- tCoupleTemp parameter
- Temperature coupling parameters
- TEMPAVG
- Standard output
- temperature parameter
- Initialization
- timestep parameter
- Timestep parameters
- TMD parameter
- Targeted Molecular Dynamics (TMD)
- TMDDiffRMSD parameter
- Targeted Molecular Dynamics (TMD)
- TMDFile parameter
- Targeted Molecular Dynamics (TMD)
- TMDFile2 parameter
- Targeted Molecular Dynamics (TMD)
- TMDFinalRMSD parameter
- Targeted Molecular Dynamics (TMD)
- TMDFirstStep parameter
- Targeted Molecular Dynamics (TMD)
- TMDInitialRMSD parameter
- Targeted Molecular Dynamics (TMD)
- TMDk parameter
- Targeted Molecular Dynamics (TMD)
- TMDLastStep parameter
- Targeted Molecular Dynamics (TMD)
- TMDOutputFreq parameter
- Targeted Molecular Dynamics (TMD)
- topology psfgen command
- List of Commands
- TOTAL2 energy
- Standard output
- TOTAL3 energy
- Standard output
- twoAwayX
- Improving Parallel Scaling
- twoAwayY
- Improving Parallel Scaling
- twoAwayZ
- Improving Parallel Scaling
- units used for output
- PDB files
| DCD trajectory files
| DCD trajectory files
| NAMD binary files
| Output files
| Standard output
- updateBias parameter
- Parameters for ABF
- useConstantArea parameter
- Tcl scripting interface and
| Pressure Control
- useConstantRatio parameter
- Tcl scripting interface and
| Pressure Control
- useFlexibleCell parameter
- Tcl scripting interface and
| Pressure Control
- useGroupPressure parameter
- Tcl scripting interface and
| Pressure Control
- useSettle parameter
- Bond constraint parameters
- vdwForceSwitching parameter
- Non-bonded force field parameters
- vdwGeometricSigma parameter
- Non-bonded force field parameters
- velDCDfile parameter
- Output files
- velDCDfreq parameter
- Output files
- velocities parameter
- Input files
- velocityQuenching parameter
- Velocity quenching parameters
- waterModel parameter
- Water Models
- wrapAll parameter
- Periodic boundary conditions
- wrapNearest parameter
- Periodic boundary conditions
- wrapWater parameter
- Periodic boundary conditions
- writenamdbin psfgen command
- List of Commands
- writepdb psfgen command
- List of Commands
- writepsf psfgen command
- List of Commands
- XSTfile parameter
- Periodic boundary conditions
- XSTfreq parameter
- Periodic boundary conditions
- zeroMomentum parameter
- Conserving momentum
http://www.ks.uiuc.edu/Research/namd/