



Next: An atom selection example
Up: Atom selections in Python
Previous: The built-in atomsel type
Contents
Index
VMD provides an atom selection class for use in the Python interpreter.
Instances of this class correspond to a set of atom indices in a
particular molecule for a particular coordinate set. Once an atom
selection is made, you can query the properties of the selected atoms,
such as their names, residue ids, or coordinates. In a similar fashion,
you can set the values of these properties. You can also perform logical
operations on atom selections, including finding the intersection or
union of two atom selections or finding the inverse of the set. Finally,
you can perform tuple operations on the atom selection object to query
the indices of the atoms in the selection.
Atom selection macros can be defined using the macro method of the
AtomSel module. The syntax is just as in the corresponding atomselect
macro and atomselect delmacro Tcl commands; see
section 9.3.2 for details.
Below we summarize the methods available from the AtomSel class.
- AtomSel(selection = 'all', molid = 0, frame = 0):
Creates a new atom selection object.
 sel = AtomSel('name CA', 1) # Selects the alpha carbons of molecule 1
sel = AtomSel('name CA', 1) # Selects the alpha carbons of molecule 1
- select(selection):
Change the selected atoms.
 sel.select('resid 5')
sel.select('resid 5')
- list():
Return a copy of the selected atom indices.
- frame(value = -1):
Set/get the coordinate frame for the selection. Nonpositive values
will return the current value of the frame without changing it.
 sel.frame(5)
sel.frame(5)
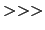 sel.frame()
sel.frame()
5
- get(attr1, attr2, ...):
Takes any number of string arguments, which should correspond to a
valid atom property such as "name", "x", or "water". Returns a list
of the value of the property for each atom in the selection. For
boolean properties such as "water", the returned value will be
1 if true and 0 if false.
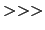 x, y, z = sel.get('x', 'y', 'z')
x, y, z = sel.get('x', 'y', 'z')
- set(attr, val):
Set the atom property corresponding to attr using the values in
val. The number of elements in val should be either 1 or
the number in the atom selection.
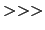 len(sel)
len(sel)
12
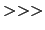 sel.set('beta',3)
sel.set('beta',3)
 sel.set('beta',(1,2,3,4,5,6,7,8,9,10,11,12))
sel.set('beta',(1,2,3,4,5,6,7,8,9,10,11,12))
- write(filename, filetype=None):
Write the atoms in the selection to filename. Filetype is guessed
from the filename, or can be specified with filetype.
- sel1 & sel2:
Create a new atom selection using the atoms found in both sel1
and sel2.
- sel1
 sel2:
Create a new atom selection using the atoms found in either sel1
or sel2.
sel2:
Create a new atom selection using the atoms found in either sel1
or sel2.
- -sel:
Create a new atom selection using the atoms not found in sel.
- len(sel):
Returns the number of atoms in the selection.
- sel[0], sel[0:3]:
Index and slice operations return the corresponding atoms in the
selection.
- center(weight=None):
Return the center of the selected atoms, possibly weighted by
weight, which must be a sequence.
- sasa(srad, samples=-1, points=None, restrict=None):
Returns the solvent-accessible surface area (SASA) of atoms in the
selection using the assigned radius for each atom, extending
each radius by srad to find the points on a sphere that are
exposed to solvent. If a restrict selection is given,
only solvent-accessible points near that selection bill be considered.
The points parameter can be used to collect the points which
are determined to be solvent-accessible; this must be a list
variable.
- getbonds():
Returns a list of the atoms bonded to each atom in the selection.
- setbonds(bonds):
Set the bonds for the atoms in the selection. bonds must be a list
of the same length as the selection; each element in the list must be
a sequence containing the indices of the atoms to which the atom has a bond.
- minmax():
Returns the minimum and maximum coordinates of the atoms in the selection
as a tuple of the form (xmin, ymin, zmin), (xmax, ymax, zmax).
- rmsd(sel, frame=None, weight=None):
Returns the root-mean-square distance of the atoms in sel from
the selection. If frame is given, the coordinates from the
corresponding frame will be used (see the example). If weight
is given, the computed RMSD will be weighted using the values in weight.
- align(ref=None, move=None, frame=None, weight=None):
Finds the transformation that aligns the atoms in the selection with the
atoms in ref, with optional weights weight, and applies this
transformation to the atoms in move. The following default values
for all arguments are provided:
- ref: Same molecule and atoms as selection, but always using the
- align(ref=None, move=None, frame=None, weight=None):
Finds the transformation that aligns the atoms in the selection with the
atoms in ref, with optional weights weight, and applies this
transformation to the atoms in move. The following default values
for all arguments are provided:
- ref: Same molecule and atoms as selection, but always using the
first timestep in the molecule.
- move: All atoms in the selection molecule.
- frame: overrides both the selection's frame and the move
frame, but does not affect the ref frame.
- weight: Defaults to uniform weights on all atoms in selection.
first timestep in the molecule.
- move: All atoms in the selection molecule.
- frame: overrides both the selection's frame and the move
frame, but does not affect the ref frame.
- weight: Defaults to uniform weights on all atoms in selection.
- contacts(cutoff, sel=None):
Returns pairs of of atoms within cutoff of each other. If sel
is None, atoms in the pairs must be in the selection; otherwise, the
first atom in each pair will be from the selection, and the second will
be from sel.




Next: An atom selection example
Up: Atom selections in Python
Previous: The built-in atomsel type
Contents
Index
vmd@ks.uiuc.edu
![]() sel = AtomSel('name CA', 1) # Selects the alpha carbons of molecule 1
sel = AtomSel('name CA', 1) # Selects the alpha carbons of molecule 1
![]() sel.select('resid 5')
sel.select('resid 5')
![]() sel.frame(5)
sel.frame(5)
![]() sel.frame()
sel.frame()
![]() x, y, z = sel.get('x', 'y', 'z')
x, y, z = sel.get('x', 'y', 'z')
![]() len(sel)
len(sel)
![]() sel.set('beta',3)
sel.set('beta',3)
![]() sel.set('beta',(1,2,3,4,5,6,7,8,9,10,11,12))
sel.set('beta',(1,2,3,4,5,6,7,8,9,10,11,12))