VMD 1.8.6
VMD 1.8.6 screen shot:
Molefacture structure editing
The Theoretical and Computational Biophysics Group is pleased to announce
VMD version 1.8.6.
VMD provides exciting new capabilities for exporting molecular graphics
to 3-D PDF files, high dynamic range images, and eerily "three dimensional"
renderings using ambient occlusion lighting with production quality
renderers.
The MacOS X and Unix versions of VMD now benefit from far more extensive use
of multiprocessor (and multicore) acceleration, greatly improving performance
for several common structure analysis operations.
By default VMD will now use all available processor cores to
accelerate parallelized portions of the code which currently include
several structure analysis routines, interactive molecular dynamics,
and ray tracing.
Many new and updated structure building
and analysis tools have been added in this release, easing
the process of setting up, running, and analyzing computer
simulations of biomolecules.
This release also contains many performance and efficiency
improvements benefiting those loading, displaying, and analyzing
large structures with millions of atoms, as well as
thousands of medium sized molecules.
Major features included in VMD 1.8.6:
Acrobat3D 3-D PDF capture
VMD 1.8.6 includes a new rendering mode which supports scene capture with
Adobe Acrobat3D,
allowing researchers to create PDF documents containing 3-D scenes from VMD.
The resulting PDF documents can be viewed on all popular computer platforms
with the free Adobe Acrobat Reader software. VMD scenes exported to 3-D
PDF files can also be imported into Microsoft PowerPoint presentations and
interactively zoomed and rotated during presentations.
A short
VMD 3-D PDF tutorial is available,
describing the 3-D capture process and suggestions to achieve good results
with the currently shipping version of Acrobat3D.
VMD now incorporates expanded support for high-quality molecular scene
rendering using
Tachyon,
NVIDIA Gelato,
and
PIXAR RenderMan.
These renderers support advanced lighting modes such as
ambient occlusion lighting, which can be used to
create molecular renderings that look more "three dimensional".
These renderers also support high dynamic range lighting (HDR) and
high precision color image formats.
A tutorial is available,
to guide users in experimenting with the use of Tachyon
ambient occlusion lighting in their VMD renderings.
Tachyon is compiled-into VMD and also supplied as a standalone program
distributed with VMD. The included Tachyon ray tracer can be used to begin
producing figures and movies using the advanced rendering features
described above without installing any additional software.

High quality GPU-accelerated
rendering with NVIDIA Gelato
One of the unique advantages of VMD's support for Gelato is the ability
to use GPUs to accelerate the rendering process while still
supporting tremendous scene complexity and retaining the high output
quality that professional software renderers are known for.
A new
Gelato tutorial is
available. The gelato tutorial describes the new Gelato export feature
and includes several early example images produced with VMD in combination
with Gelato.
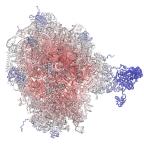
Time-averaged trajectory
Coulombic potential map

Electrostatic field lines
representation
VMD includes a new "Field Lines" representation which can be used
to display images of electrostatic fields, flow fields, and
other volumetric data for which viewing field lines or
particle advection traces is helpful.
The "volmap" command in VMD now provides a Coulombic potential map
averaging function which can be used to process molecular dynamics
trajectories, producing a time-averaged potential field which can be
used for focused NAMD simulations on substructures within a larger simulation.
VMD outline rendering style
While not yet a standardized rendering feature, users can now easily
experiment with non-photorealistic shading in VMD 1.8.6.
A new
tutorial and example scenes
show how to create VMD renderings
which attempt to imitate the style of David S. Goodsell,
famous for his paintings and artistic renderings of molecules.
The example scene and tutorial override the standard VMD GLSL shading
feature with a non-photorealistic shader that adds adjustable
edge enhancement outlines which can be faded in and out with the
standard diffuse shading of VMD's GLSL rendering mode.
VMD 1.8.6 uses just over half as much memory relative to prior versions,
allowing structures of up to 72 million atoms to be loaded and displayed
on workstations with 16GB of memory, using the 64-bit versions of VMD.
The reduction in memory use similarly
benefits researchers loading thousands of structures at a time.
With adequate physical memory (22GB), a single text-mode instance of VMD can
load and operate on the entire PDB databank (30,000+ molecules) at once.
While an extreme example, this illustrates what is now possible as a result
of these efficiency improvements.
VMD 1.8.6 is the first version of VMD to include support for
GPU acceleration of non-graphical computations.
VMD can be compiled to use
NVIDIA CUDA
to accelerate calculation of Coulombic potential maps produced by the
new volmap "coulomb" map type. Since CUDA itself is still in beta and
functionality and implementation details may change, VMD CUDA
support is currently limited to source-based compilations. We will
release fully CUDA-enabled binaries of VMD concurrently with the official
NVIDIA CUDA product announcement. Subsequent releases of VMD will benefit
from more extensive application of GPU acceleration to several of the most
time consuming trajectory analysis commands in VMD.
Full list of newly added features and improvements in VMD 1.8.6