From: Francesco Pietra (chiendarret_at_gmail.com)
Date: Thu Mar 01 2018 - 03:05:22 CST
Hi Brian:
I went on with that FEP of previous thread "Ligand atoms moving too fast
with FEP" protein-ligand complex. Aim: estimating the binding free energy
with respect to other ligands for the same binding site.
frwd Bound was run with delta lambda 0.2, i.e., five windows, with 100,000
steps re-equilibration and 400,000 steps FEP at each window, ts=1.0fs.
Is it possible guessing from parseFEP, from simply frwd, if such conditions
lead to acceptable convergence? Or back running is also needed for such a
guess? Or, finally, should be better running frwd, or forwd+back, with ten
or more windows?
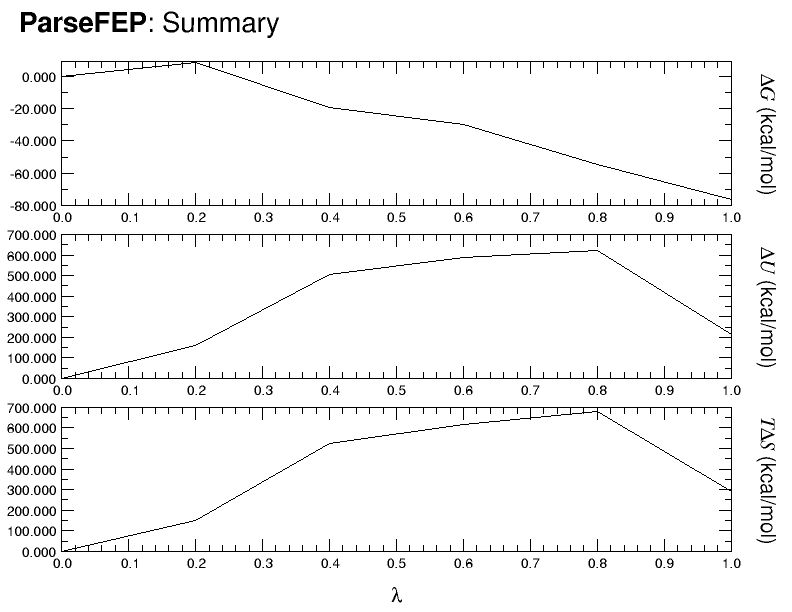
>From the attached summary.png, from parseFEP, for the above frwd, I fear
that the above conditions for frwd are inadequate (convergence far from
being attained) to my purpose. All such questions because I am dealing with
a large system, with costly simulations.
Thanks a lot for advice.
francesco pietra

This archive was generated by hypermail 2.1.6 : Mon Dec 31 2018 - 23:20:53 CST