From: Ajasja Ljubetič (ajasja.ljubetic_at_gmail.com)
Date: Mon Nov 17 2014 - 10:28:20 CST
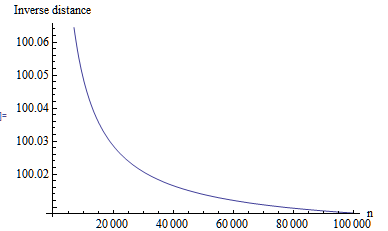
Here is a quick plot of inverse distance versus n for a uniform
distribution on the interval (1,2). As you can see, with increasing n the
value of the distance approximates the minimum distance.
[image: Inline images 1]
If your distances are large, you may have to set n to large values.
So barring any precision issues this approach should work.
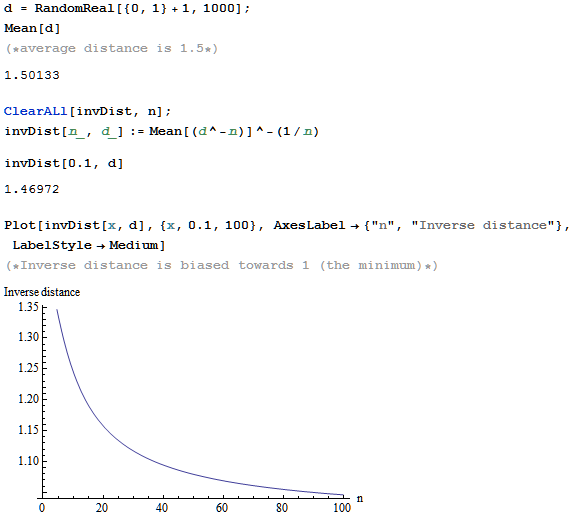
For example for values in the range of (100,101) with n=10000
[image: Inline images 2]
Good luck & best regards,
Ajasja
On 17 November 2014 17:14, Vlad Cojocaru <vlad.cojocaru_at_mpi-muenster.mpg.de>
wrote:
> Below is how PLUMED implements the minimal distance and this looks more
> in the direction you mention ..... The problem with PLUMED is that in
> apparently does not allow NPT simulations with NAMD ... I also don't know
> how PMF calculations with PLUMED versus COLVAR scale with NAMD
>
> http://plumed.github.io/doc-v2.0/user-doc/html/mindist.html
>
> Best
> Vlad
>
>
>
> On 11/17/2014 05:07 PM, Jérôme Hénin wrote:
>
> Indeed, but the larger the negative exponent, the more this average is
> skewed towards the smallest distances in the set, making it a possible
> approximation to the minimal distance. If that is not steep enough, the
> power function can be replaced with an exponential, but it's still a form
> of average. By the way, that would be really easy to implement if there is
> demand for it.
>
> Jerome
>
> On 17 November 2014 16:57, Vlad Cojocaru <
> vlad.cojocaru_at_mpi-muenster.mpg.de> wrote:
>
>> But this looks more like an average distance than a minimum distance,
>> isn't it ?
>>
>>
>>
>> On 11/17/2014 03:49 PM, Giacomo Fiorin wrote:
>>
>> Can you try distanceInv choosing the exponent that gives you the best
>> behavior?
>> On Nov 17, 2014 8:42 AM, "Vlad Cojocaru" <
>> vlad.cojocaru_at_mpi-muenster.mpg.de> wrote:
>>
>>> Dear NAMD users,
>>>
>>> Could you please let me know if it is currently possible to define the
>>> minimal distance between 2 groups of atoms as a colvar component within the
>>> colvar module ?
>>>
>>> Thanks
>>>
>>> Best wishes
>>> Vlad
>>>
>>> --
>>> Dr. Vlad Cojocaru
>>> Computational Structural Biology Laboratory
>>> Department of Cell and Developmental Biology
>>> Max Planck Institute for Molecular Biomedicine
>>> Röntgenstrasse 20, 48149 Münster, Germany
>>> Tel: +49-251-70365-324; Fax: +49-251-70365-399
>>> Email: vlad.cojocaru[at]mpi-muenster.mpg.de
>>> http://www.mpi-muenster.mpg.de/research/teams/groups/rgcojocaru
>>>
>>>
>> --
>> Dr. Vlad Cojocaru
>> Computational Structural Biology Laboratory
>> Department of Cell and Developmental Biology
>> Max Planck Institute for Molecular Biomedicine
>> Röntgenstrasse 20, 48149 Münster, Germany
>> Tel: +49-251-70365-324; Fax: +49-251-70365-399
>> Email: vlad.cojocaru[at]mpi-muenster.mpg.dehttp://www.mpi-muenster.mpg.de/research/teams/groups/rgcojocaru
>>
>>
>
> --
> Dr. Vlad Cojocaru
> Computational Structural Biology Laboratory
> Department of Cell and Developmental Biology
> Max Planck Institute for Molecular Biomedicine
> Röntgenstrasse 20, 48149 Münster, Germany
> Tel: +49-251-70365-324; Fax: +49-251-70365-399
> Email: vlad.cojocaru[at]mpi-muenster.mpg.dehttp://www.mpi-muenster.mpg.de/research/teams/groups/rgcojocaru
>
>


This archive was generated by hypermail 2.1.6 : Wed Dec 31 2014 - 23:23:01 CST