|
|
JMV: Java Molecular Viewer |
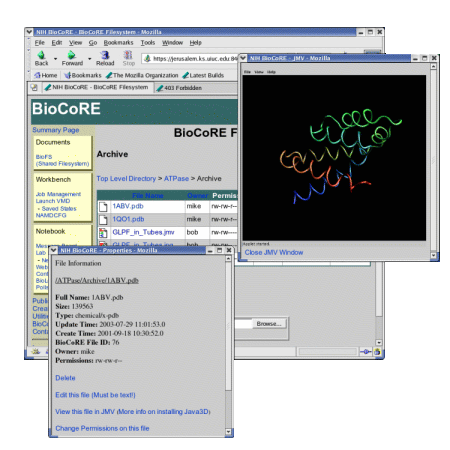
JMV is a Java applet that displays views of molecules from a web browser equipped with Java3D. JMV will work on many versions of Unix and Windows, unlike web-based molecular viewers implemented as Netscape plug-ins. JMV provides different representations, such as Tube, Trace, and CPK, for a molecule and can view PDB files from both the BioFS and the Protein Data Bank. JMV is written in a modular fashion using Java Beans and is one of a number of tools that will be available outside as well as inside BioCoRE. As a standalone tool, JMV can be used on a web page to provide a rotatable, scalable view of a molecule.
|
|
While working, Bob uses a PDB file, a type of data file that describes a molecule. He loads this file into the BioFS for others to view. The file can be viewed from within BioCoRE with JMV, the Java Molecular Viewer. In the BioFS, Bob opens the file in JMV, and the molecule is displayed. He can then view the molecule, move it and change the representation and color. Once he gets it into a state that that shows an important feature, he saves a state file back to the BioFS, where others can see it.
|
 (Click here for a larger, more detailed view)
(Click here for a larger, more detailed view)
|
|
|
|
|
