Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
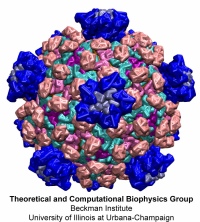
image size:
219.5KB
made with VMD
Viruses are the cause of many human diseases, from the common cold to
AIDS, and medicine is continuously searching for better ways to
battle viruses through vaccination or medication. Detailed knowledge
of the life cycles of viruses should be useful in the treatment of
viral diseases. A key focus of investigations is the virus capsid, a
protein coat that protects the viral genome, but also triggers
release of the genome and other viral factors upon contact with the
body's cells. X-ray crystallography has resolved the average
structures of many types of virus capsids, providing the basis for
detailed investigations, for example by means of molecular dynamics
methods, of capsid dynamical properties, e.g., in assembly and
disassembly. Unfortunately, due to their large size most virus
capsids are beyond the reach of molecular dynamics simulations, with
one notable exception
(see the March 2006
highlight "Simulating an Entire Life Form"). This earlier simulation
allowed researchers to develop and test a method for coarse-grained
molecular dynamics simulations that glosses over atomic detail and,
thereby, permits microsecond descriptions of entire viral particles. As reported recently
(see also journal cover)
such simulations, employing the program NAMD,
were applied to the empty capsids of several viruses. These simulations
revealed a variety of behaviors, from rapid collapse to high stability,
depending on the strength of interactions between the proteins from which
capsids are built. The new method offers unprecedented views of capsid
dynamics that may assist in battling viral diseases. More information on
the simulations can be found on our virus web page.



