Nandan Haloi
I am a Ph.D. student at the center for Biophysics and Quantitative Biology at the University of Illinois at Urbana-Champaign (UIUC). My research interest involves structural modeling of proteins and studying protein dynamics with the aim of developing new medicines. I am currently working on two major projects:
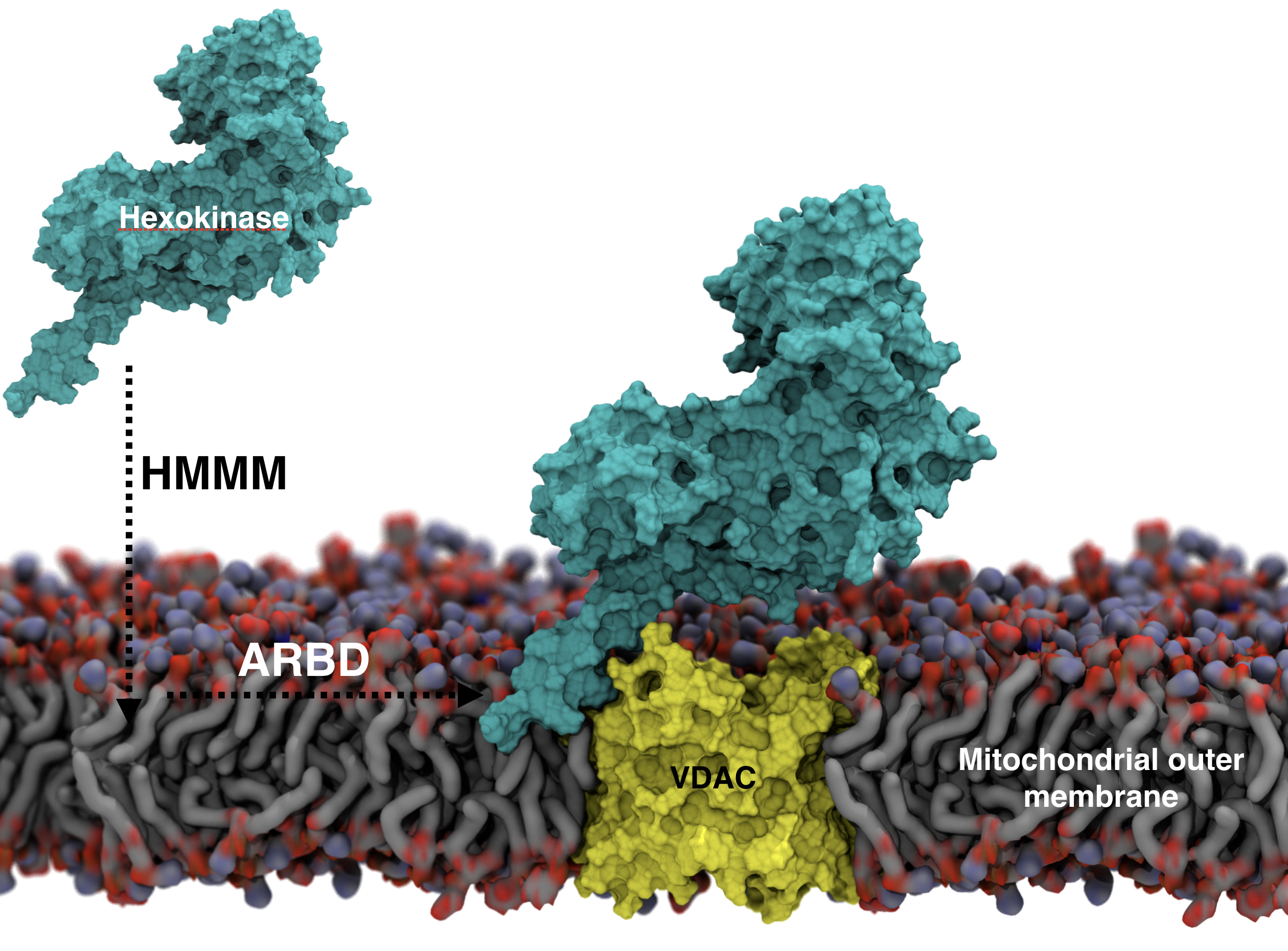
1) Studying the membrane binding of hexokinase (HK) and followed by its complex formation with membrane-embedded VDAC at the surface outer membrane of mitochondrial. Complex formation between HK and the mitochondrial channel VDAC plays a crucial role in regulating cell growth and survival; however, structural details of this complex remain elusive. We used molecular dynamics combined with Brownian dynamics simulations and proposed a novel structural model of the complex between membrane-bound HK with VDAC. Our model provided mechanistic insight into the complex formation mechanism. The model was further verified by site-directed mutagenesis, conducted by the groups of Dr. Wai-Meng Kwok and Dr. Amadou Camara at Wisconsin Medical School. (manuscript under preparation)
2) Studying antibiotic permeation through bacterial outer membrane porins using molecular dynamics simulations. Developing new antibiotics against Gram-negative bacteria is a huge challenge as drugs face additional barriers crossing the lipopolysaccharide-coated outer membrane. However, antibiotics in the presence of a primary amine group can easily permeate through porins present in the outer membrane, discovered recently by our experimental collaborator, Dr. Paul J. Hergenrother's group at UIUC. However, the reason behind enhanced permeation was not well understood. We are using molecular docking coupled with free energy calculation methods to characterize the permeation mechanism of newly synthesized antibiotics by our collaborator and trying to delineate the reason behind the enhanced permeation.
Home Department: Biophysics and Quantitative Biology
Office Address: Beckman Institute for Advanced Science and Technology, Room 3017, 405 N Mathews Ave, Urbana, IL 61801
Email Address: nhaloi2@illinois.edu
Education
- B.Tech. in Chemical Science and Technology, Indian Institute of Technology, Guwahati (India), 2012-2016 (group Dr. Debasis Manna)
Internships
- Summer research internship at École polytechnique fédérale de Lausanne, Switzerland with Dr. Matteo Dal Peraro, 2019
- Summer research internship at Saint Louis University, USA with Dr. Ryan D McCulla, 2015
Skills
- Experimental lab techniques: Protein purifications, Fluorescence spectroscopy, GC-MS, GC-FID
- Molecular dynamics techniques: BEUS, Force field development, HMMM, ARBD, SMD
- Software packages: NAMD, VMD, Gaussian09, AutoDock, HADDOCK2.2
- Programming skills: Tcl, Bash and Python. Good knowledge in Linux environment
Research Interests
- Cryo-EM structure modeling.
- Drug design.
- Protein-lipid and protein-protein interactions.


Publications during B.Tech
Gorai, S.; Paul, D.; Haloi, N.; Borah, R.; Santra, M. K.; Manna, D. Molecular BioSystems, 2016, 12:747-757
Gorai, S.; Paul, D.; Borah, R.; Haloi, N.; Santra, M. K.; Manna, D. Chemistry Select, 2018, 3(4):1205-1214




