



Next: Exercise 3: The
physical
Up: Residue Selection
Previous: Exercise 1:
Investigating Structure Contents
Before beginning this exercise, change the highlighting style back to Bonds
and the Molecule Coloring to Sequence Identity per residue.
In order to display sequence conservation, follow these steps.
- 1 Go back to the main MultiSeq window top pull-down
menu and select Tools
 Residue Selection. Residue Selection.
- 2 Select Sequence Identity.
- 3 In the Sequence Identity window, set the measure
to Greater than.
- 4 Select the value 0.5.
- 5 Click the Select button.
|
 |
Do you notice a difference in the highlighted areas, in comparing the
structure (Q) to sequence? Upon examining the residues in the Sequence
display, notice how the majority of the sequence conservation occurs
among the top 3 molecules. Since the top 3 molecules are
AspRS and the bottom is SerRS, this pattern of sequence conservation
makes sense.
Go back to the Sequence Identity window. Decrease the value to 0.2 and
click Select. Take note of how many conserved residues are in
the OpenGL Display and Sequence Display. Now, increase the
value back to 0.5. You will see, as you progress, the yellow color
diminishing among the molecules.
Figure 16:
Sequence Identity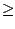 0.7
0.7

|
The yellow coloring remains towards the core of the molecule and at the
dimerization site. Increase the value to 0.7 and click Select;
you should notice that the only highlighted sequences in the Sequence
Display are those that match exactly. Finally, change the value to 1.0.
Between Sequence Identity 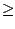 0.7 and Sequence
Identity
0.7 and Sequence
Identity 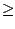 1.0, there are only 10 residues which are
identical and span all four molecules. Go back to the Sequence Identity
window and click on the Close Window button.
1.0, there are only 10 residues which are
identical and span all four molecules. Go back to the Sequence Identity
window and click on the Close Window button.
Figure 17:
Highlighted residues in Sequence Display

|




Next: Exercise 3: The
physical
Up: Residue Selection
Previous: Exercise 1:
Investigating Structure Contents
Brijeet Dhaliwal
2004-09-15









