

Next: Solvating the Protein
Up: Basics of NAMD
Previous: What is Needed
Of the four files mentioned above, an initial pdb file will typically be obtained through the Protein Data Bank, and the parameter and topology files for a given class of molecule may be obtained via the Internet at
Notice that the latest release (c31b1) also contains the updated CMAP correction, used in this tutorial. The psf file must be created by the user from the initial pdb and topology files. The NAMD configuration file is also created by the user, with commands based on the specific requirements for the MD simulation.
- 1
- Go to the 1-1-build directory. In a Terminal window, you can change directories using the cd command. Type cd 1-1-build. You can see the contents of the directory you are in by typing ls. In this folder, you will find many files that you will use later.
First, you will remove the water molecules from 1UBQ.pdb, and create a pdb file of the protein alone.
- 2
- Open VMD by typing vmd in the Terminal window.
namd-tutorial-files/1-1-build> vmd
|
|
- 3
- Load 1UBQ.pdb by clicking File
 New Molecule... menu item in the VMD Main window. In the Molecule File Browser use the Browse... button to find the file 1UBQ.pdb. Load it by pressing the Load button.
New Molecule... menu item in the VMD Main window. In the Molecule File Browser use the Browse... button to find the file 1UBQ.pdb. Load it by pressing the Load button.
Note that the X-ray structure from the Protein Data Bank does not contain the hydrogen atoms of ubiquitin. This is because X-ray crystallography usually cannot resolve hydrogen atoms. The pdb file you will generate with psfgen along with the psf will contain guessed coordinates for hydrogen atoms of the structure. Later, energy minimization of the protein will ensure their positions are reasonable.
- 4
- Choose the Extensions
 Tk Console menu item and in
the VMD TkCon window. Be sure you are in the 1-1-build directory. If you are not, navigate there using the ls command to list files and directories and the cd command to change directories. Then, type the following commands:
Tk Console menu item and in
the VMD TkCon window. Be sure you are in the 1-1-build directory. If you are not, navigate there using the ls command to list files and directories and the cd command to change directories. Then, type the following commands:
| set ubq [atomselect top protein] |
|
$ubq writepdb ubqp.pdb
|
|
(Hit the Return key after each command.)
In the previous step you have created the file ubqp.pdb, which contains the coordinates of the ubiquitin alone without hydrogens, in the 1-1-build directory.
- 5
- Quit VMD by choosing File
 Quit.
Quit.
- 6
- Now, you will create the psf file of ubiquitin. Note that VMD offers an automatic psf file builder via the VMD Main menu by clicking Extensions
 Modeling
Modeling  Automatic PSF Builder. We will create the psf file manually to teach you exactly how it is done. The psfgen package of VMD is very useful in this regard. In order to create a psf, you will first make a pgn file, which will be the target of psfgen.
Automatic PSF Builder. We will create the psf file manually to teach you exactly how it is done. The psfgen package of VMD is very useful in this regard. In order to create a psf, you will first make a pgn file, which will be the target of psfgen.
In a Terminal window type VMD text editor to open the text editor.
Type in the following lines:
| package require psfgen |
|
| topology top_all27_prot_lipid.inp |
|
| pdbalias residue HIS HSE |
|
| pdbalias atom ILE CD1 CD |
|
| segment U {pdb ubqp.pdb} |
|
| coordpdb ubqp.pdb U |
|
| guesscoord |
|
| writepdb ubq.pdb |
|
writepsf ubq.psf
|
|
- 7
- After typing this, save the file by clicking File
 Save. Be sure that you are in the 1-1-build directory and enter the
file name as ubq.pgn. Quit the text editor by clicking File
Save. Be sure that you are in the 1-1-build directory and enter the
file name as ubq.pgn. Quit the text editor by clicking File  Exit.
Exit.
The file you just created contains the necessary commands to create the psf file of ubiquitin with hydrogen atoms and without water. Each command of the pgn file is explained:
- Line 1: You will be running psfgen within VMD. This line requires that the psfgen package is available to be called by VMD.
- Line 2: Load the topology file top_all27_prot_lipid.inp
- Line 3: Change the residue name of histidine to the proper name found in the topology file. HSE is one of three names for histidine, based on the protonation state of its side group. See the science box below for more information.
- Line 4: The atom named ``CD1" (
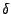 carbon) in isoleucine residues is renamed as ``CD", its proper name from the topology file. Since isoleucine contains only one
carbon) in isoleucine residues is renamed as ``CD", its proper name from the topology file. Since isoleucine contains only one 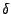 carbon atom, the psf file does not use the number label after ``CD".
carbon atom, the psf file does not use the number label after ``CD".
- Line 5: A segment called U is created, containing all atoms from ubqp.pdb. The segment command also adds hydrogen atoms.
- Line 6: Coordinates are read from ubqp.pdb and residue and atom names are matched. Old segment labels will be overridden with the new segment label ``U".
- Line 7: Coordinates of missing atoms (like hydrogens) are guessed based on residue definitions from the topology file.
- Line 8: A new pdb file with the complete coordinates of all atoms, including hydrogens, is written.
- Line 9: A psf file with the complete structural information of the protein is written.
- 8
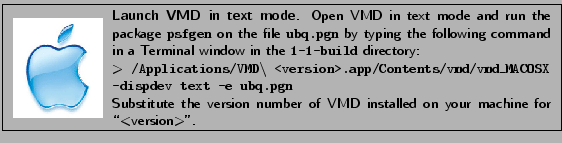
- In a Terminal window (again, be sure that you are in the 1-1-build directory), type the following command:
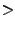 vmd -dispdev text -e ubq.pgn vmd -dispdev text -e ubq.pgn
|
|
This will run the package psfgen on the file ubq.pgn and generate the psf and the pdb file of ubiquitin with hydrogens.
In your screen you will see different messages. Warnings are related to the ends of your molecule and are normal. Your system should have 1231 atoms and 631 with guessed coordinates.
- 9
- Type exit in the Terminal to exit VMD.
Having run psfgen, two new files will now appear in your 1-1-build directory: ubq.pdb and ubq.psf. Check this by typing ls in a Terminal. You have created the psf! You may want to inspect ubq.pdb using VMD text editor .
Figure 2:
Flowchart indicating the role of files as used by VMD, NAMD, and psfgen.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{pictures/tut_unit01_flow}
}
\end{center}
\end{figure}](img24.png) |


Next: Solvating the Protein
Up: Basics of NAMD
Previous: What is Needed
namd@ks.uiuc.edu
![% latex2html id marker 3756
\framebox[\textwidth]{
\begin{minipage}{.2\textwid...
...certain model peptides. More in Appendix \ref{ap-paramfiles}.}
\end{minipage} }](img16.png)
![% latex2html id marker 3756
\framebox[\textwidth]{
\begin{minipage}{.2\textwid...
...certain model peptides. More in Appendix \ref{ap-paramfiles}.}
\end{minipage} }](img16.png)



![\framebox[\textwidth]{
\begin{minipage}{.2\textwidth}
\includegraphics[width=2...
...\tt psfgen} package requires a {\tt pdb} file for each chain.}
\end{minipage} }](img23.png)