

Next: Setup
Up: Stretching Deca-Alanine
Previous: Stretching Deca-Alanine
Introduction
In this session, you will be introduced to interactive molecular
dynamics (IMD) and steered molecular dynamics (SMD) simulations, and
to the calculation of potential of mean force (PMF) from trajectories
obtained with SMD simulations.
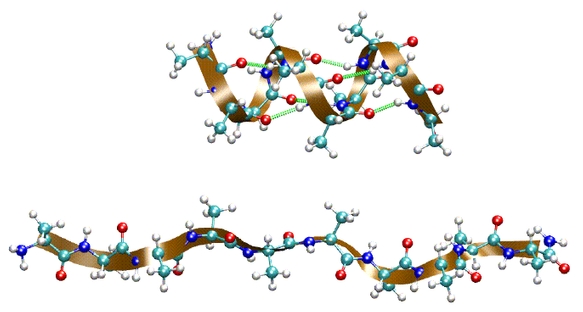
You will be using one system throughout: deca-alanine. Deca-alanine is
a peptide composed of ten alanine residues. You will simulate it in
vacuum. In vacuum deca-alanine forms an alpha-helix. That is, the
alpha-helix is the stable conformation of the molecule in vacuum, as
opposed to the beta-strand or the random coil. The helix is shown in
the top figure. It is stabilized by six hydrogen bonds (shown in
green).
Using IMD and SMD, you will stretch the molecule by applying an
external force. As the molecule is stretched, it will undergo a
gradual conformational change from the alpha-helix to the random coil
(bottom figure). Using SMD trajectories and employing Jarzynski's
equality, we will calculate the PMF involved in the helix-coil
transition.


Next: Setup
Up: Stretching Deca-Alanine
Previous: Stretching Deca-Alanine

