

Next: SMD simulation
Up: Stretching Deca-Alanine
Previous: Setup
IMD simulation
Let's run an IMD simulation of deca-alanine. Yes, you will be
interacting with the simulation.
1. Starting an IMD simulation
Files needed:
da.psf -- protein structure
imd_ini.pdb -- initial coordinates
par_all27_prot_lipid.prm -- CHARMM parameters
imd.namd -- NAMD configuration
imdfixedatoms.pdb -- fixed atoms
The following part of imd.namd enables IMD (already in the
file):
IMDon on
IMDport 3000 ;# port number (enter it in VMD)
IMDfreq 1 ;# send every 1 frame
IMDwait yes ;# wait for VMD to connect before running?
Run NAMD:
tbss> namd2 imd.namd >! da_imd.log &
The simulation will not actually run until the connection between NAMD
and VMD is established.
2. VMD control of an IMD simulation
Start VMD:
tbss> vmd -e imd.vmd
This will load the molecule and show it in both the Ribbon and the CPK
representations, as saved in the VMD state file imd.vmd. You
will see one atom (alpha-carbon of the first residue) colored in
orange. That atom will be fixed during the IMD simulation.
Within VMD, select the menu item Extensions -> imd .
In the IMD Connection window, enter Hostname:
localhost and Port: 3000
Click Connect.
You should see the molecule jiggling as the simulation is running.
Even though the double representation of Ribbon and CPK is good for
viewing the overall and detailed structure of the molecule, you may
change the representation to whatever you want. You can change the
representation while the IMD simulation is running.
3. Applying a force in an IMD simulation
|
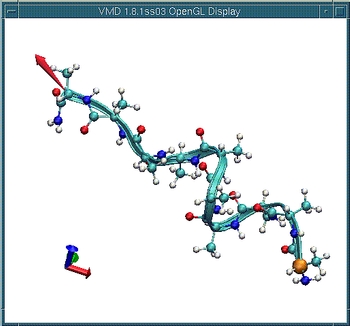
Now let's apply a force to stretch the molecule.
Select the menu item Mouse -> Force -> Atom.
Click and drag on an atom located near the other side of the `orange'
atom. The red arrows indicates the force being applied (magnitude and
direction). Stretch the molecule, say, by half of its original
length.
Now remove the force by middle-clicking on the atom to which the force
is being applied.
Observe how the molecule folds back.
| |
 |
If you want to stop the simulation, click Stop Sim in the
IMD Connection window. Otherwise, the simulation is set to run
for 100 ps (about 10-20 minutes of your time). Try various things:
pull different atoms, squash the molecule, do whatever you want. Use
your imagination.
When the simulation stops, the molecule will stop jiggling.
Quit VMD (the menu item File -> Quit).
If you want to run the IMD simulation again, repeat the procedure of
this section starting with running NAMD.
Later in the Summer School, you will be introduced to a VMD extension
called AutoIMD, which is another way of doing IMD simulations. It
automatically generates all the necessary files and launch and connect
to a NAMD simulation.


Next: SMD simulation
Up: Stretching Deca-Alanine
Previous: Setup
