



Next: Moving frame of reference.
Up: Selecting atoms
Previous: Selecting atoms
Contents
Index
Selection keywords may be used individually or in combination with each other, and each can be repeated any number of times.
Selection is incremental: each keyword adds the corresponding atoms to the selection, so that different sets of atoms can be combined.
However, atoms included by multiple keywords are only counted once.
Below is an example configuration for an atom group called ``atoms''.
Note: this is an unusually varied combination of selection keywords, demonstrating how they can be combined together: most simulations only use one of them.
atoms {
# add atoms 1 and 3 to this group (note: the first atom in the system is 1)
atomNumbers {
1 3
}
# add atoms starting from 20 up to and including 50
atomNumbersRange 20-50
# add all the atoms with occupancy 2 in the file atoms.pdb
atomsFile atoms.pdb
atomsCol O
atomsColValue 2.0
# add all the C-alphas within residues 11 to 20 of segments "PR1" and "PR2"
psfSegID PR1 PR2
atomNameResidueRange CA 11-20
atomNameResidueRange CA 11-20
# add index group (requires a .ndx file to be provided globally)
indexGroup Water
}
The resulting selection includes atoms 1 and 3, those between 20 and 50, the
 atoms between residues 11 and 20 of the two segments PR1 and PR2, and those in the index group called ``Water''.
The indices of this group are read from the file provided by the global keyword indexFile.
atoms between residues 11 and 20 of the two segments PR1 and PR2, and those in the index group called ``Water''.
The indices of this group are read from the file provided by the global keyword indexFile.
In the current version, the Colvars module does not manipulate VMD atom selections directly: however, these can be converted to atom groups within the Colvars configuration string, using selection keywords such as atomNumbers.
The complete list of selection keywords available in VMD is:
-
atomNumbers
 List of atom numbers
List of atom numbers
Context: atom group
Acceptable values: space-separated list of positive integers
Description: This option adds to the group all the atoms whose numbers are in
the list. The number of the first atom in the system is 1: to convert from a VMD selection, use ``atomselect get serial''.
-
indexGroup
 Name of index group to be used (GROMACS format)
Name of index group to be used (GROMACS format)
Context: atom group
Acceptable values: string
Description: If the name of an index file has been provided by indexFile, this option allows to select one index group from that file: the atoms from that index group will be used to define the current group.
-
atomsOfGroup
 Name of group defined previously
Name of group defined previously
Context: atom group
Acceptable values: string
Description: Refers to a group defined previously using its user-defined name.
This adds all atoms of that named group to the current group.
-
atomNumbersRange
 Atoms within a number range
Atoms within a number range
Context: atom group
Acceptable values:  Starting number
Starting number -
- Ending number
Ending number
Description: This option includes in the group all atoms whose numbers are within the range specified. The number of the first atom in the system is 1.
-
atomNameResidueRange
 Named atoms within a range of residue numbers
Named atoms within a range of residue numbers
Context: atom group
Acceptable values:  Atom name
Atom name
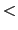 Starting residue
Starting residue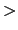 -
-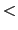 Ending residue
Ending residue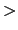
Description: This option adds to the group all the atoms with the provided
name, within residues in the given range.
-
psfSegID
 PSF segment identifier
PSF segment identifier
Context: atom group
Acceptable values: space-separated list of strings (max 4 characters)
Description: This option sets the PSF segment identifier for
atomNameResidueRange. Multiple values may be provided,
which correspond to multiple instances of
atomNameResidueRange, in order of their occurrence.
This option is only necessary if a PSF topology file is used.
-
atomsFile
 PDB file name for atom selection
PDB file name for atom selection
Context: atom group
Acceptable values: UNIX filename
Description: This option selects atoms from the PDB file provided and adds them
to the group according to numerical flags in the column
atomsCol. Note: the sequence of atoms in the PDB file
provided must match that in the system's topology.
-
atomsCol
 PDB column to use for atom selection flags
PDB column to use for atom selection flags
Context: atom group
Acceptable values: O, B, X, Y, or Z
Description: This option specifies which PDB column in atomsFile is used to determine which atoms are to be included in the group.
-
atomsColValue
 Atom selection flag in the PDB column
Atom selection flag in the PDB column
Context: atom group
Acceptable values: positive decimal
Description: If defined, this value in atomsCol identifies atoms in atomsFile that are included in the group.
If undefined, all atoms with a non-zero value in atomsCol are included.
-
dummyAtom
 Dummy atom position (Å)
Dummy atom position (Å)
Context: atom group
Acceptable values: (x, y, z) triplet
Description: Instead of selecting any atom, this option makes the group a virtual particle at a fixed position in space. This is useful e.g. to replace a group's center of geometry with a user-defined position.




Next: Moving frame of reference.
Up: Selecting atoms
Previous: Selecting atoms
Contents
Index
vmd@ks.uiuc.edu
![]() atoms between residues 11 and 20 of the two segments PR1 and PR2, and those in the index group called ``Water''.
The indices of this group are read from the file provided by the global keyword indexFile.
atoms between residues 11 and 20 of the two segments PR1 and PR2, and those in the index group called ``Water''.
The indices of this group are read from the file provided by the global keyword indexFile.