Known Issue: The output of hbonds cannot be considered 100% accurate if the donor and acceptor selection share a common set of atoms.
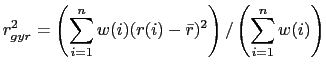 |
(9.1) |
where
The following options can be specified:
- molid <default molid>: The default molecule to which an atom belongs unless a molecule number was explicitely specified for this atom in the atom list. Further, all frame specifications refer to this molecule. (Default is the current top molecule.)
- frame <frame>: By default the value for the current frame will be returned but a specific frame can be chosen through this option. One can also specify all or last instead of a frame number in order to get a list of values for all frames or just the last frame respectively.
- first <frame>: Use this option to specify the first frame of a frame range. (Default is the current frame.)
- last <frame>: Use this option to specify the last frame of a frame range. (Default is the last frame of the molecule).
Here are a few examples of usage:
measure bond {3 5} - Returns the distance between atoms 3 and 5 of the
current frame of the top molecule. measure bond {3 5} molid 1 frame all - Returns the distance between
atoms 3 and 5 of molecule 1 for all frames.
measure bond {3 {5 1}} molid 0 first 7 - Returns the distance between
atoms 3 of molecule 0 and atom 5 of molecule 1. The value is computed for all
frames between the seventh and the last frame of molecule 0.
- k <value>: force constant for bond, angle, dihed and imprp energies in
kcal/mol/A
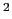 or kcal/mol/rad
or kcal/mol/rad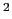 respectively.
respectively.
- x0 <value>: equilibrium value for bond length, angle, dihedral angles and improper dihedrals in Angstrom or degree.
- kub <value>: Urey-Bradley force constant for angles in kcal/mol/A
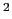 .
.
- s0 <value>: Urey-Bradley equilibrium distance for angles in Angstrom.
- n <value>: dihedral periodicity.
- delta <value>: dihedral phase shift in degree (usually 0.0 or 180.0).
- rmin1 <value>: VDW equilibrium distance for atom 1 in Angstrom.
- rmin2 <value>: VDW equilibrium distance for atom 2 in Angstrom.
- eps1 <value>: VDW energy well depth (epsilon) for atom 1 in kcal/mol.
- eps2 <value>: VDW energy well depth (epsilon) for atom 2 in kcal/mol.
- q1 <value>: charge for atom 1.
- q2 <value>: charge for atom 2.
- cutoff <value>: nonbonded cutoff distance.
- switchdist <value>: nonbonded switching distance.
Here is a 2D example of a nonorthogonal PBC cell: A and B are the are the displacement vectors which are needed to create the neighboring images. The parallelogram denotes the PBC cell with the origin O at its center. The square to the right indicates the orthonormal unit cell i.e. the area into which the atoms will be wrapped by transformation T.
+ B
/ + B'
_________/________ |
/ / / +---|---+
/ / / T | | |
/ O--------/-------> A ====> | O---|--> A'
/ / | |
/_________________/ +-------+
A = displacement vector along X-axis with length a
B = displacement vector in XY-plane with length b
A' = displacement vector along X-axis with length 1
B' = displacement vector along Y-axis with length 1
O = origin of the PBC cell
|
The second argument (cutoff) is the maximum distance (in Å) from the PBC unit cell for atoms to be considered. In other words the cutoff vector defines the region surrounding the pbc cell for which image atoms shall be constructed (i.e. {6 8 0} means 6 Å for the direction of A, 8 Å for B and no images in the C-direction).
The following options can be specified:
- molid <molecule_number>: The default molecule to which an atom belongs unless a molecule number was explicitely specified for this atom in the atom list. Further, all frame specifications refer to this molecule. (Default is the current top molecule.)
- frame <frame>: By default the value for the current frame will be returned but a specific frame can be chosen through this option. One can also specify all or last instead of a frame number in order to get a list of values for all frames or just the last frame respectively.
- sel <selection>: If an atomselection is provided then only those image atoms are returned that are within cutoff of the selected atoms of the main cell. In case cutoff is a vector the largest value will be used.
- align <matrix>: In case the molecule was aligned you can supply the alignment matrix which is then used to correct for the rotation and shift of the pbc cell.
- boundingbox PBC
 {<mincoord> <maxcoord>}: With this option the atoms are
wrapped into a rectangular bounding box. If you provide "PBC" as an argument then the
bounding box encloses the PBC box but then the cutoff is added to the bounding box.
Negative values for the cutoff dimensions are allowed and lead to a smaller box.
Instead you can also provide a custom bounding box in form of the minmax coordinates
(list containing two coordinate vectors such as returned by the measure minmax command).
Here, again, the cutoff is added to the bounding box.
{<mincoord> <maxcoord>}: With this option the atoms are
wrapped into a rectangular bounding box. If you provide "PBC" as an argument then the
bounding box encloses the PBC box but then the cutoff is added to the bounding box.
Negative values for the cutoff dimensions are allowed and lead to a smaller box.
Instead you can also provide a custom bounding box in form of the minmax coordinates
(list containing two coordinate vectors such as returned by the measure minmax command).
Here, again, the cutoff is added to the bounding box.
Options:
- tol <value>: Allows one to control tolerance of the algorithm when considering wether something is symmetric or not. A smaller value signifies a lower tolerance, the default is 0.1.
- nobonds: If this flag is set then the bond order and orientation are not considered when comparing structures.
- verbose <level>: Controls the amount of console output. A level of 0 means no output, 1 gives some statistics at the end of the search (default). Level 2 gives additional info about each stage, and 2, 3, 4 yield even more info for each iteration.
- idealsel <selection>: The symmetry search will be performed on the regular selection but then the found symmetry elements will be imposed on the selection given with this option an the search is repeated with this second selection. This method allows, for example, to perform the symmetry guess on a selection without hydrogens (which might point in random directions for rotable groups) but still get the ideal coordinates and unique atoms for the entire structure. The selection specified here must be a superset of the selection used for the symmetry search.
- I: Instead of guessing the symmetry pointgroup of the selection determine if the selection's center off mass represents an inversion center. The returned value is a score between 0 and 1 where 1 denotes a perfect match.
- plane <vector>: Instead of guessing the symmetry pointgroup of the selection determine if the plane with the defined by its normal vector is a mirror plane of the selection. The returned value is a score between 0 and 1 where 1 denotes a perfect match.
- Cn
 Sn <vector>:
Instead of guessing the symmetry pointgroup of the selection
determine if the rotation or rotary reflection axis Cn/Sn
with order n defined by vector exists for the
selection. E.g., if you want to query wether the Y-axis
has a C3 rotational symmetry you specify C3 {0 1 0}.
The returned value is a score between 0 and 1 where 1
denotes a perfect match.
Sn <vector>:
Instead of guessing the symmetry pointgroup of the selection
determine if the rotation or rotary reflection axis Cn/Sn
with order n defined by vector exists for the
selection. E.g., if you want to query wether the Y-axis
has a C3 rotational symmetry you specify C3 {0 1 0}.
The returned value is a score between 0 and 1 where 1
denotes a perfect match.
- imposeinversion: Impose an inversion center on the structure.
- imposeplanes {<vector> [<vector> ...]}: Impose the planes given by a list of normal vectors on the structure.
- imposeaxes|imposerotref {<vector> order [<vector> order ...]}:
Impose rotary axes or rotary reflections on the structure specified by a list of pairs of a vector and an integer. Each pair defines an axis and its order.
Result:
The return value is a TCL list of pairs consisting of a label string and a value or list. For each label the data following it are described below:
- pointgroup
- The guessed point group. For point groups that have an order associated with it, like C3v or D2, the order is replaced by 'n' and we have Cnv or Dn. The order is given separately (see below).
- order
- Point group order, i.e. order of highest axis (0 if not applicable).
- elements
- Summary of found symmetry elements, i.e. inversion center, rotary axes, rotary reflections, mirror planes. Example: ``(i) (C3) 3*(C2) (S6) 3*(sigma)'' for point group D3d.
- missing
- Elements missing with respect to ideal set of elements (same format as above). If this is not an empty list then something has gone awfully wrong with the symmetry finding algorithm.
- additional
- Additional elements that would not be expected for this point group (same format as above). If this is not an empty list then something has gone awfully wrong with the symmetry finding algorithm.
- com
- Center of mass of the selection based on the idealized coordinates (see 'ideal' below).
- inertia
- List of the three axes of inertia, the eigenvalues of the moments of inertia tensor and a list of three 0/1 flags specifying for each axis wether it is unique or not.
- inversion
- Flag 0/1 signifying if there is an inversion center.
- axes
- Normalized vectors defining rotary axes
- rotreflect
- Normalized vectors defining rotary reflections
- planes
- Normalized vectors defining mirror planes.
- ideal
- Idealized symmetric coordinates for all atoms of the selection. The coordinates are listed in the order of increasing atom indices (same order asa returned by the atomselect command ``get x y z''). Thus you can use the list to set the atoms of your selection to the ideal coordinates (see example below).
- unique
- Index list defining a set of atoms with unique coordinates.
- orient
- Matrix that aligns molecule with GAMESS standard orientation.
If a certain item is not present (e.g. no planes or no axes) then the corresponding value is an empty list. The pair format allows to use the result as a TCL array for convenient access of the different return items.
Example:
set sel [atomselect top all]
# Determine the symmetry
set result [measure symmetry $sel]
# Create array 'symm' containing the results
array set symm $result
# Print selected elements of the array
puts $symm(pointgroup)
puts $symm(order)
puts $symm(elements)
puts $symm(axes)
# Set atoms of selection to ideally symmetric coordinates
$sel set {x y z} $symm(ideal)
Classify the space surrounding a molecule as interior or exterior. For structures not representing a closed container, or in applications where interior and exterior boundaries are ill-defined such as surface cleft identification, fuzzy boundary detection should be enabled with the -probmap flag.
When measure volinterior is invoked, a QuickSurf density of the atom selection is first computed; the -res and -isovalue parameters correspond to QuickSurf parameters used in generating the molecular density. For each voxel not corresponding to molecular density (i.e., selection voxels), rays are cast in parallel. In the default, fixed boundary implementation: if a ray strikes the system boundary before selection voxels, then that voxel is considered exterior and is marked accordingly and ignored for the remainder of the calculation. A Tcl list is returned with four entries: total voxel count, exterior voxel count, interior voxel count, and selection voxel count. The resulting volume loaded into -mol molid is a discrete 3D grid, where each voxel is classified as interior, exterior, or selection.
For the fuzzy boundary implementation, i.e. the -probmap flag is given, all
![]() rays are cast and the number of rays striking selection voxels before the
system boundary is stored for each voxel.
The continuous grid output by the fuzzy boundary implementation is normalized
against
rays are cast and the number of rays striking selection voxels before the
system boundary is stored for each voxel.
The continuous grid output by the fuzzy boundary implementation is normalized
against ![]() rays such that all entries vary continuously on
rays such that all entries vary continuously on ![]() .
The directions of rays are randomly chosen and are uniformly distributed on the
surface of the unit sphere following a Poisson disk sampling procedure, although
uniformity of ray directions below -nrays 32 cannot be guaranteed.
Thus, values passed to -nrays of at least 32 are recommended if the
-probmap flag is given. In addition to the continuous 3D grid loaded into
-mol molid, a Tcl list is returned containing ten entries, where each
entry is the number of voxels in the grid at percentiles ranging from 10-99th.
Optionally, the -discretize flag will process the continuous grid into a
discrete grid, equivalent to that output by the fixed boundary implementation,
according to the user-specified cutoff value. If -discretize is given,
then a Tcl list of total, exterior, interior and selection voxel counts is
returned.
.
The directions of rays are randomly chosen and are uniformly distributed on the
surface of the unit sphere following a Poisson disk sampling procedure, although
uniformity of ray directions below -nrays 32 cannot be guaranteed.
Thus, values passed to -nrays of at least 32 are recommended if the
-probmap flag is given. In addition to the continuous 3D grid loaded into
-mol molid, a Tcl list is returned containing ten entries, where each
entry is the number of voxels in the grid at percentiles ranging from 10-99th.
Optionally, the -discretize flag will process the continuous grid into a
discrete grid, equivalent to that output by the fixed boundary implementation,
according to the user-specified cutoff value. If -discretize is given,
then a Tcl list of total, exterior, interior and selection voxel counts is
returned.
For more information, please refer to the publication [47]:
https://doi.org/10.1021/acs.jcim.9b00324
Options:
- nrays <value> The number of rays to cast from each voxel. A minimum of six rays is required.
- spacing <value> Grid spacing parameter, in units of Å. This parameter impacts performance, but must also be considered when converting voxel counts into physical volumes.
- res <value> QuickSurf resolution, in units of Å.
Default value is
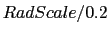 , where
RadScale corresponds to the radius scale value used when generating the
QuickSurf density.
, where
RadScale corresponds to the radius scale value used when generating the
QuickSurf density.
- isovalue <value> Isovalue to use when creating the QuickSurf density.
- mol <molid> Molecule into which the resulting volumetric data are loaded. If this flag is not set, voxel counts are returned but no volumetric data are loaded.
- vol <volid> Specify an existing target volume by its volume ID.
- probmap Enable fuzzy boundary detection. This is necessary if the
molecule does not represent a closed container with a continuous surface,
or a closed container but with a porous surface.
If this flag is enabled, the resulting volume
grid stores entries varying continuously on
![$ [0,1]$](img72.png) , where a value of 1.0
corresponds to 100% probability of that voxel being interior. A Tcl list
of 10 values is also returned, with voxel counts given for each percentile
ranging from 10th to 99th percentile.
, where a value of 1.0
corresponds to 100% probability of that voxel being interior. A Tcl list
of 10 values is also returned, with voxel counts given for each percentile
ranging from 10th to 99th percentile.
- discretize <cutoff> Create a discrete grid from the continuous probability grid using the specified cutoff and load it into molecule molid. This option is valid only if -probmap has been specified. The cutoff value must be between 0 and 1.0, and is used to generate a discrete volume grid equivalent to that output from the fixed boundary, implementation. With this flag set, voxel counts are returned in the same manner as the fixed-boundary implementation, i.e. as a Tcl list of four values: total voxel count, interior voxel count, exterior voxel count, selection voxel count.
- verbose Enable verbose printing.
- overwrite <volid> Overwrite volume volid already loaded into molecule molid. If no volume is loaded corresponding to volid, an error is thrown.
Fixed boundary example:
set id [mol new myMolecule.pdb]
set sel [atomselect $id "noh protein"]
set result [measure volinterior $sel -nrays 12 \
-isovalue 1.0 -spacing 1.0 -res [expr 1.2/0.2] \
-mol $id -verbose]
set totalVoxels [lindex $result 0]
set exteriorVoxels [lindex $result 1]
set interiorVoxels [lindex $result 2]
set selectionVoxels [lindex $result 3]
# Use output volume grid to make a selection
set watersInterior [atomselect $id "water and vol0 < 1 and vol0 > -1"]
Fuzzy boundary example:
set id [mol new myMolecule.pdb]
set sel [atomselect $id "noh protein"]
# Continuous grid output
set resultC [measure volinterior $sel -nrays 64 \
-isovalue 0.8 -spacing 1.0 -res [expr 1.0/0.2] \
-mol $id -verbose -probmap]
set 50th_percentile [lindex $resultC 4]
set 60th_percentile [lindex $resultC 5]
# Use output to select waters in regions above 85% likelihood of being interior
set watersInterior [atomselect $id "water and vol0 > 0.85"]
# Discrete grid output
set resultD [measure volinterior $sel -nrays 64 \
-isovalue 0.8 -spacing 1.0 -res [expr 1.0/0.2] \
-mol $id -verbose -probmap -discretize 0.85]
set totalVoxels [lindex $resultD 0]
set exteriorVoxels [lindex $resultD 1]
set interiorVoxels [lindex $resultD 2]
set selectionVoxels [lindex $resultD 3]
# Use discretized grid to select interior waters,
# equivalent to above selection on continuous grid.
## Note that we now use vol1, because we did not choose to overwrite the
## previous volume (vol0).
set watersInterior2 [atomselect $id "water and vol1 < 1 and vol1 > -1"]