If no selection is given, the macro for the given name is returned.
If no name is given, a list of all macro names is returned.
If a macro already exists for the given name, the old selection will be replaced with the new selection. Singlewords that are not defined as macros, like protein and water, cannot be redefined with the macro command.
Some examples are:
vmd> atomselect top "name CA" atomselect0 vmd> atomselect 3 "resid 25" frame last atomselect1 vmd> atomselect top "within 5 of resname LYR" frame 23 atomselect2
The newly created atom selection is a Tcl proc, which takes the following options:
- num: Return the number of atoms in the selection.
- list: Return a list of the atom indices in the selection (BTW, this is the same as get index).
- text: Return the text used to create this selection.
- molid: Returns the molecule id used to create this selection.
- frame: Returns the animation frame associated with this selection. The
result will be either now, last, or an integer corresponding
to the frame. When the frame is now, the atom selection will use
atomic coordinates from the current frame for its associated molecule. If
the frame is last, the atom selection will always use coordinates
from the last frame. If the frame is a specific integer, the selection
will always use coordinates from that frame, even if the current animation
frame changes. Note that if a nonexistent frame is specified, the atomic
coordinates will all be taken to be
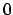 .
.
- frame frame: Set the frame for the selection. frame should be either now, last, or an integer.
- delete: Delete this object (removes the function).
- global: Moves the object into the global namespace. Atom selections created within a Tcl proc that are not made global are deleted when the proc exits.
- uplevel level: Moves the object to a new level in the namespace stack. Works the same as the Tcl function uplevel.
- get attribute_list: Given an attribute or a list of
attributes, returns the attribute values. If only a single attribute is
given, a list of corresponding attributes values will be returned. If a
list of attributes is given, then a list of sublists will be returned;
each sublist will contain the values for the corresponding attributes.
See Tables 5.5 and 5.6 for the
recognized attribute keywords.
- set attribute_list values_lists: Set the attributes
in the attribute list with the values gven in the values lists.
If there is only one attribute, then values_lists can be either
a single value or a list of values, one for each selected atom.
If there is more than one attribute, then values_lists must be a
list of sublists; the number of sublists must equal the number of selected
atoms, and the number of items in each sublist must equal the number of
attributes.
Example:
set sel [atomselect top all] set mass [$sel get mass] set xyz [$sel get {x y z}] $sel set beta 0 # all values are set to zero $sel set beta $mass # copy mass to beta # set occupancy to x, mass to y, beta to z $sel set {occupancy mass beta} $xyzIt is an error to set integer or floating point keywords using non-numeric values. If floating point values are passed to integer keywords, they will be converted to integers, and vice versa.
The set command immediately updates all representations of the selected molecule. If speed is an issue, delete all representations of the molecule before setting the values.
- getbonds: returns a list of bondlists; each bondlist contains the id's of the atoms bonded to the corresponding atom in the selection.
- setbonds: Set the bonds for the atoms in the selection; the second argument should be a list of bondlists, one bondlist for each selected atom.
- move 4x4 matrix: Applies the given transformation matrix to the coordinates of each atom in the selection.
- moveby offset: move all the atoms by a given offset.
- lmoveby offset_list: move each atom by an offset given in the list.
- moveto position: move all the atoms to a given location.
- lmoveto position_list: move each atom to a point given by the appropriate list element.
- writeXXX filename: write the selected atoms to a file of type XXX; e.g., pdb, dcd. New in VMD 1.8: writepdb requires a filename; omitting the filename no longers returns the PDB data as a string. To get the PDB data as a string, first write to a file, then enter the following commands: set fd [open filename r]; set s [read $fd]; close $fd. The text will be contained in the variable s.
- update: Update the atom selection based on the frame for the
selection (the frame can be specified using the frame option as
described above).
See section 11.2 for more on using atom selections for fun and profit, as well as issues relating to speed of analysis scripts.