From: Lara rajam (lara.4884_at_gmail.com)
Date: Tue Sep 01 2015 - 12:12:42 CDT
Dear NAMD !
I have already posted this mail , If I can get some help, that will be
useful for me !
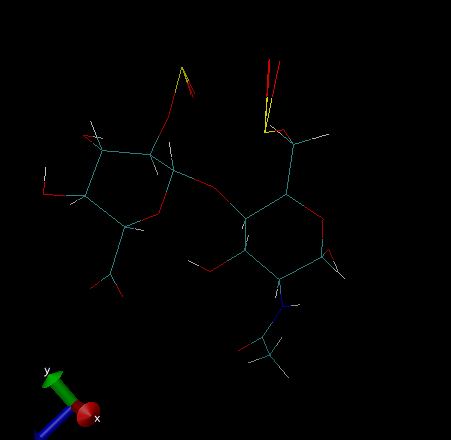
I am trying to do simulation of heparin disaccharide ( iduronic acid 2
sulfate and nacetyle glucosamine 6 sulfate)
I got the topology file top_all36_carb.rtf and prm files
I built it using vmd and used patch to add glycosidic linkage
I solvated and added counter ions. when I started the minimization and
equilibration the SO3 group is disturbed and all the oxygens got merged
I have attached the image for reference(removed solvent molecules). could
any one suggest me how to fix this issue
thank you

This archive was generated by hypermail 2.1.6 : Thu Dec 31 2015 - 23:22:02 CST