From: nicolas martin (nicolasmartin973_at_gmail.com)
Date: Thu May 22 2014 - 07:06:08 CDT
Please do not take into account the first attachment but this one. The
first one was a mistake.
2014-05-22 13:21 GMT+02:00 nicolas martin <nicolasmartin973_at_gmail.com>:
> Dear all, drear Giacomo,
>
> As you advised me, I decided to move on with a new molecule,
> ethyl-benzene. Same as before concerning my purpose. I define here the
> rotation axis for the spinangle as the bond between the atom on the ring
> and the next to it which is the first of the ethyl moiety. So I compute the
> free energy of the rotation of the final carbon (my selection is actually
> final carbon the one before and the 2 H on it) of the ethyl group around
> the bound defined beforehand. I must admit that I have no more fitting
> problems in VMD so I guess no more in the refgroup fitting if they are both
> using the same algorithm.
>
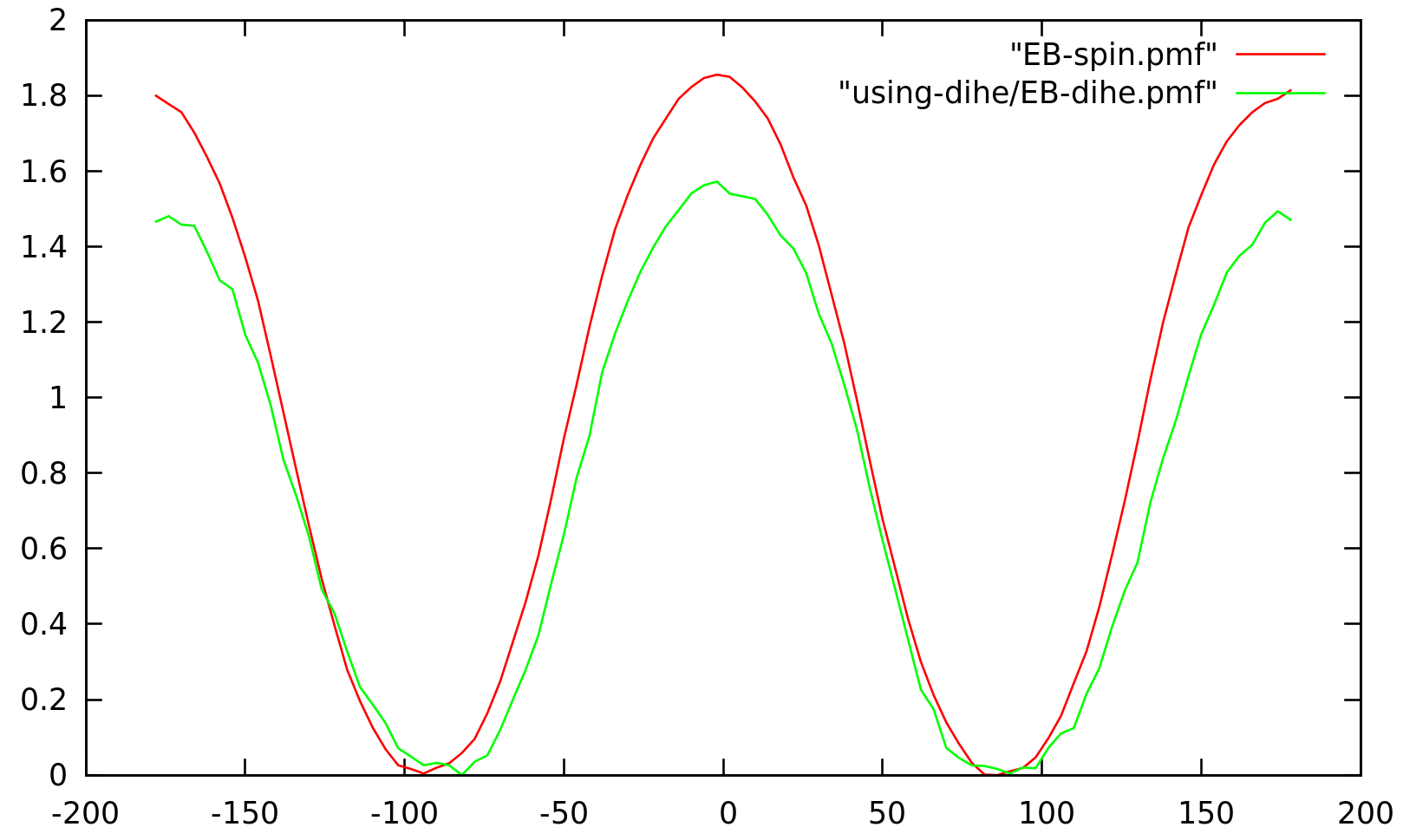
> But I came up with a new problem. I can observe a shift on the PMF (see
> file attached) between both methods I used, dihedral and spinangle. If I'm
> right, the way I set everything should lead me to the same profile. But I'm
> concerned with one detail. Since the center of geometry of my selection to
> compute the spinangle (the last part of my ethyl moeity) is not exactly on
> the axis defined beforehand, the axis will be first shifted to go through
> this center of geometry and might this somehow interact with the rotation
> energy ? I have checked the applied forces and can't get anything relevant
> for my problem out of it. Any ideas?
>
> Thanks a lot for you help
>
>
>
>
> 2014-05-12 0:27 GMT+02:00 Giacomo Fiorin <giacomo.fiorin_at_gmail.com>:
>
> Hello Nicolas, as I mentioned earlier and Jérôme pointed out as well,
>> butane is a very small and flexible molecule, not suitable for defining a
>> rigid-body rotation. Including the hydrogens won't help, because they are
>> even more flexible than the carbon atoms, in particular the methyl
>> hydrogens which can rotate freely around the C1-C2 bond. (I wrote to you
>> about this too).
>>
>> In short, NAMD can simulate butane just fine, but your assumption that
>> you can have a large enough rigid section (more than 3 atoms) is incorrect.
>>
>> Please, do try with a molecule that includes a rigid group like a benzyl
>> ring. (See my previous email).
>>
>> Thank you
>> Giacomo
>>
>>
>> On Sun, May 11, 2014 at 5:52 PM, nicolas martin <
>> nicolasmartin973_at_gmail.com> wrote:
>>
>>> Hi Jérôme,
>>>
>>> Thank you for your answer. I tried to include in my refgroup selection
>>> the hydrogens around C1 and C2. So the selection contains now 8 atoms. But
>>> still, the result remains the same.
>>>
>>> Do you think skipping this step assuming that it is due to the fact that
>>> butane is not really what NAMD is made to work with and going to the next
>>> step which is actual work on my protein would be a good idea? I mean how
>>> can I make sure that I am not mistaken there and that it’s just a matter of
>>> size and not because of me not fully understanding how to use that
>>> powerful tool ?
>>>
>>> Thank you again, bests,
>>>
>>> NM
>>>
>>>
>>> 2014-05-09 20:53 GMT+02:00 Jérôme Hénin <jerome.henin_at_ibpc.fr>:
>>>
>>> Hi Nicolas,
>>>>
>>>> I think the most likely issue is that 3 atoms is not sufficient for the
>>>> refPositionsGroup to be fitted properly. With this setup you should be able
>>>> to make it a little larger.
>>>>
>>>> Cheers,
>>>> J
>>>>
>>>> ----- Original Message -----
>>>> > Dear NAMD users,
>>>> >
>>>> > I ask for your help once more concerning my issues with spinangle.
>>>> > I'm working on butane (I know this is not ideal due to the fact that
>>>> it's a
>>>> > small molecule that could create problems for fittings) and I'm
>>>> trying to
>>>> > use spinangle covlar to compute a PMF along the rotation of the
>>>> dihedral
>>>> > angle defined by C1 C2 C3 and C4. This is done on purpose because I
>>>> plan to
>>>> > move on a protein so the obviously solution of using a dihedral colvar
>>>> > isn't my first choice. C1 C2 and C3 are included into the reference
>>>> group
>>>> > and if I'm right this allows me to cancel any rotation or translation
>>>> > effects to THEN compute the spinangle with contains as well a fitting
>>>> > between a reference structure a the current one (superimposed with the
>>>> > atoms specified in refgroup as said before).
>>>> >
>>>> > I tried first the following colvar defnition and in addition used the
>>>> > keyword fixedAtoms to fix C1 C2 and C3 and I got a nice match between
>>>> this
>>>> > PMF and the one obtained with a simple dihedral colvar (my reference).
>>>> >
>>>> > Then I removed the fixedAtoms keyword and here is my problem, my PMF
>>>> is not
>>>> > anymore what I expected.
>>>> >
>>>> > So please tell me where is my mistake? I thought it would be the same
>>>> and I
>>>> > wouldn't need to fix any atoms since I'm fitting C1 C2 and C3 to the
>>>> > reference structure...?
>>>> >
>>>> > And if I move on my protein, how can I then make sure than the
>>>> diffusion
>>>> > (even if it shoul be really less than butane's diffusion) isn't
>>>> introducing
>>>> > a bias in my PMF?
>>>> >
>>>> >
>>>> > Thanks a lot in advance for your help ,
>>>> >
>>>> > NM
>>>> >
>>>> >
>>>> > spinangle.in :
>>>> >
>>>> > colvarsTrajFrequency 1
>>>> >
>>>> > colvar {
>>>> > name spinangle
>>>> > spinangle {
>>>> > axis (-1.408,-0.537,-0.317) # axis of bond C2 C3
>>>> >
>>>> > atoms {
>>>> > atomNumbers 8 9 10 11 #..moving group
>>>> > centerReference on
>>>> > rotateReference on
>>>> > refPositionsGroup { #..reference group
>>>> > atomNumbers 1 5 8
>>>> > }
>>>> > refPositionsFile ../input/buta.pdb
>>>> > }
>>>> > refPositionsFile ../input/buta.pdb
>>>> > }
>>>> > }
>>>> >
>>>> >
>>>> >
>>>> > harmonic {
>>>> > name harmonic
>>>> > colvars spinangle
>>>> > forceConstant 0.006
>>>> > centers XXXX
>>>> > }
>>>> >
>>>>
>>>
>>>
>>
>

This archive was generated by hypermail 2.1.6 : Wed Dec 31 2014 - 23:22:25 CST